Agent-Based Model: A Surging Tool to Simulate Infectious Diseases in the Immune System ()
Keywords: Agent-Based Model; Complex Biological Processes; Progression of Infectious Disease
1. Introduction
Infectious disease, identified by clinical symptoms, is defined as the presence and growth of a various type of pathogen in an organism [1]. Under most circumstances, intruding pathogens are eliminated by activating immune cells such as tissue macrophages and activated neutrophils in the immune system. If overwhelming immune response occurs, unbalanced responses between immune cells and cytokines lead to unexpected harmful outcomes for patients. Abundant research has recently focused on modeling immune responses to infectious disease such as sepsis or gut infection in order to explore complicated dynamic presentation of cells and cytokines in the immune system under the presence of infection. Modeling and simulation of immune responses to infectious disease could provide dynamic understanding of infectious disease progression and further acknowledge therapeutic targets for the infectious disease.
As a standard approach, mathematical modeling is currently being developed as a dynamic knowledge representation offering a promising possibility for understanding complex local and global dynamics of infectious disease [2,3]. Using a series of known and hypothesized kinetics of biologic system components from current literature, mathematical models describe infectious disease processes by measuring the steady states of various components in the immune system. However, mathematical models fail to capture inhomogeneous information of various components over the simulation space and fail to describe possible deviations of various components from their aggregated behaviors. As a powerful computational modeling technique, Agent-based model (ABM) simulates complicated non-linear dynamic relationships between components and intuitively maps a more realistic biological system by incorporating spatial effects and stochastic nature into model construction. Key elements of ABM include agents, a collection of decision-making entities classified into different types based on entities described in a real-world system. Each type of agents executes certain behaviors appropriate for the system they represent. By implementing a pre-defined set of rules, agents move in a certain direction and arbitrarily interact with other agents in a spatial environment. Agent behaviors are updated in various locations according to update rules executed at discrete time steps. Agent-based modeling inherently captures repetitive spatial interactions between agents in a stochastic process and, therefore, is a powerful tool to render valuable information and redraw an overall picture of a biological system. Even simple implementation of ABM requires well-established technology which relies on the power of computers to explore dynamics beyond the reach of pure mathematical methods [4,5]. Because of the inherent nature in computational structure, the agent-based model can be implemented on parallel computers very efficiently [6]. This review specifically investigates previous applications of agent-based modeling to infectious disease associated with failure of the immune system to respond to intruding bacteria. Subsequent article sections are organized as follows: 1) introduction of basic structure of agent-based model, 2) review of existing research delineating ABM implementations on infectious disease, 3) discussion of the advantages and disadvantages of ABM on modeling of infectious disease, and 4) prediction of future implementation of ABM on infectious disease and other types of disease in a broader way.
2. Basic Structure of Agent-Based Model
A typical type of agent-based model includes two elements: “Agents” and “Environment”. Agents are main components of agent-based model and their interacting behaviors following local rules on known mechanism, which represent the mechanism of entire system. For instance, a typical prey-predator agent-based model involves two types of agents: preys and predators. The environment in prey-predator agent-based model is defined as a user-specified 5 by 5 squares, and each square represents specific location of simulation environment. Predators make decision to kill preys in a natural way, and therefore the number of preys will dramatically decrease if the number of predators increases within a certain range of appearance of preys. In addition, the number of preys also relies on “environment”, which is defined as food source for preys. Each square associated with food resource, in prey-predator agent-based model, containing local information of the amount of available food resource. The localized information will change the motion of both preys and predators. Preys at location k will make their own decision (which is defined by modeler in prey-predator agent-based model) to move to location j (assume location j has sufficient food resource) if location k has run out of food. Thus, the number of preys and predators will differ upon on spatial locations (available food resource) in prey-predator agent-based model. The number of preys at location k is in relation to the number of predators and available food resource at that location, referring to a specific ordinary differential mathematical model [7]. The following Figure 1 delineates a basic structure of prey-predator agent-based model.
Figure 1 shows preys at location k are assigned probability PRk,i to move to location i, probability PRk,m to move to loction m, probability PRk,j to move to location j, and PRk,n to move to location n. At the same time,
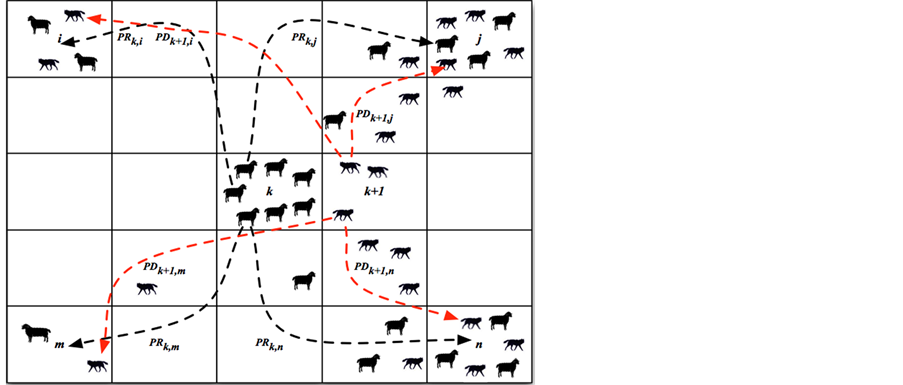
Figure 1. Basic structure of agent-based model using prey-predator interaction as an example.
predators at location k + 1 are assigned probability PDk+1,j to move to location j, PDk+1,n to move to location n, PDk+1,i to move to location i, and PDk+1,m to move to location m. It is observed most preys are moving toward location j and location n for more food resource at those two sites based on assumption, and therefore, more predators moving afterwards to location j and location n for hunting preys. By implementing prey-predator agent-based model, one can plot the progression of preys and predators over time.
3. Agent-Based Model of Sepsis
Sepsis, currently defined as a systemic inflammatory response in the presence of an infectious agent or trauma, is increasingly considered an exaggerated, poorly regulated, innate immune response to microbial products [8,9]. Progression to severe sepsis is marked by generalized hypotension, tissue hypoxia, and coagulation abnormality [10]. Severe sepsis can further develop into septic shock if long-lasting severe hypotension occurs [10] and ultimately lead to death. The first application of agent-based modeling of sepsis is employed by An [11]. An has produced a very abstract ABM of Acute Inflammatory Response, an initial stage of sepsis progression. His model is built on the interface between endothelial cells and blood at the capillary level to simulate behaviors of circulating neutrophils and monocytes in the presence of injury. Neutrophils and monocytes are defined as agents and their behaviors, including rolling, sticking, diapedesis and respiratory burst, are regulated by a series of state variables which obey fundamental occurrence in AIR environment derived from literature. Figure 2 shows interacting behaviors of macrophages, neutrophils and red blood cells in An’s agent-based model.
In Figure 2, each square represents specific intercellular location near the site of infection in An’s agent-based model. The macrophages and neutrophils are recruited from blood vessel to the infecting location based on known biological mechanism. State variables in relation to macrophages and neutrophils are defined to vary from location to location when agent-based simulation executes. The accumulation in macrophage and neutrophil-dependent state variables is used to calibrate global variables such as “total oxy deficit” and “End Injury Vector Number”. The variable “total oxy deficit” measures total damage caused by AIR and the variable “End Injury Vector Number” measures accumulated infection load during AIR progression in order to reflect the characteristics of AIR progression. Using a predefined rule system, multiple independent computer programs are executed with various initial injury extents to generate three general outcomes of AIR progression, including heal, SIRS, and overwhelming infection. A large amount of simulation data, such as the time-dependent number of neutrophils and macrophages, is collected during agent-based model implementation. Furthermore, the author generates a distributed outcome of AIR progression by calibrating the “oxy” and “End Injury Vector Number” in 500 selected iterations of simulation runs under the same extent of injury. Distribution outcomes confirm that AIR progression is stochastically represented and simulates heterogeneity of a patient population. At the end of his study, An concluded that his agent-based model could not represent a real system but is helpful for understanding essential steps in the inflammatory process at the level of his proposed model. For future research, he expects to produce simulated results that can be validated based on existing experimental studies and use more sophisticated ABMs to test therapies prior to clinical trials in order to refine clinical study design in pharmacological research.

Figure 2. Interacting behaviors among macrophages, neutrophils, and red blood cells.
Following his previous work, An continued ABM construction to simulate and compare different therapeutic effects on the improvement of patients’ outcomes [12]. The model is developed at the cellular level and, as in the previous model, built on endothelial-blood interface. Compared to the previous model, however, he incorporated additional agents to represent sophistic interaction between cells and pathways of immune responses in AIR progression. Positive/negative feedback relationships and interactions between agents are represented and updated using simple arithmetic relationships guided by cellular/molecular mechanisms of AIR progression. The range of initial injury which generates SIR becomes the zone of interest. Distributions of a variable “end oxy deficit (EOD) with respect to different initial injury levels in infectious and sterile models with and without antibiotics, demonstrated that survivability of patients would improve by using antibiotics. Furthermore, An generated four recognizable dynamic behavior patterns of infection, including healing, immune-compromised SIRS, hyper inflammatory SIR, and overwhelming infection under different levels of initial injury. In the end, he intended to test and compare effects of various sets of anti-cytokine therapies which originate from existing clinical trials, animal study and proposed interventions using the proposed agent-based model. Mortality rates associated with anti-cytokine therapies for a group of “patients” demonstrated that anti-cytokine sets are not statistically significant in regards to outcome improvement given design parameters of the clinical trials. Failure of the initial clinical trials, the author concluded, is because redundant pathways of innate immune response could cause therapy interventions to fail to hit the targeted pathway. He criticized the proposed agent-based model for being very abstract model, qualitatively calibrated and difficult to apply in clinics at the current level. More specific analysis, such as grouping septic patients, could account for mortality rates for specific groups of patients instead of a global mortality rate for patients as a whole. Furthermore, as a future research goal, quantitative agent-based models are expected to calibrate patterns of inflammatory responses using basic scientific data in order to reproduce and illustrate effects of clinical interventions.
Recently, Wu et al. proposed an integrated ABM embedded with a mathematical model to simulate AIR progression occurring at the interface between blood vessels and cells within the tissue [13]. Five of the total agents are defined in the model: pathogen, resting neutrophils, activated neutrophils, damaged tissue and anti-inflammatory cytokines. The agents’ aggregated behaviors reflected characteristics of a class of cells or cytokines in AIR progression and provided biological insight into a series of immune response processes in AIR by describing intercellular interactions among the cells and cytokines. Interactions between agents obey fundamental immune response processes in the AIR environment derived from the literature, and change in the level of each type of agent is derived from ordinary differential equations. By implementing the ABM with corresponding initial profiles of the patients of interest and adjustable system parameters, behaviors of the agents and local intercellular interactions are captured by the simulated results. These results showed three different scenarios of AIR under various combinations of initial conditions: healthy response with low pathogen load, severe sepsis and persistent non-infectious inflammations. By analyzing outputs of patients (combined levels of pathogen, resting neutrophils, activated neutrophils, damaged tissue and anti-inflammatory cytokines) with variation in different initial conditions, the authors concluded that variations in initial levels of pathogen, initial levels of anti-inflammatory cytokines, system parameters associated with anti-inflammatory cytokines, as well as system parameters associated with the pathogen, primarily influenced outputs of patients. The advantage of Wu’s agent-based model, compared to other agent-based models is to incorporate a dynamic mathematical matching to recognized biological kinetics of AIR. However, experimental data incorporation as well as experimental validation is still under development.
Other than modeling interactions between cells, Dong et al. proposed an ABM framework to model intracellular dynamics of the NF-kB signaling module and further illustrate subsequent intercellular interactions among macrophages and T-helper cells through up-regulation of inflammatory mediators [14]. Their approach explored hypothetical scenarios of AIR and potentially improved understanding of behaviors of the molecular species which could develop and expand to emergent behavior of the overall AIR system. Simulated results include five different scenarios under various initial conditions: a self-limited response where the inflammatory stimulus was cleared, a persistent infectious response where the inflammatory stimulus such as LPS failed to be eliminated, a persistent non-infectious inflammatory response where the inflammatory stimulus was eliminated but the inflammatory response was elevated by high concentration of inflammatory stimulus, and two other scenarios associated with endotoxin tolerance and potentiation effects. The advantage of this agent-based model is integration of intracellular responses among inflammatory mediators followed by intercellular responses among immune cells. The disadvantage of this model is that it still uses a qualitative measurement of AIR and does not include experimental validation.
4. Agent-Based Model for Other Types of Infectious Disease
Sepsis, or acute inflammatory response, is one kind of infectious disease of primary focus in healthcare and is used to explore immune responses to other types of infectious disease. Along with sepsis, we concluded that most infectious diseases are induced by a series of unbalanced immune responses in the immune system. Agentbased models play an essential role in building interactions between immune responses and gains insight into the unbalanced infectious disease progression. In 2007, Mi et al., proposed an agent-based model to simulate underlying biological pathways, including interactions among macrophages, neutrophils, and fibroblasts and release of cytokines such as TNF-α and TGF-β1, as a cohesive whole, in diabetic foot ulcers (DFU) while also suggesting novel therapeutic approaches for treating DFU [15]. The authors tested and proved that elevated TNF-α or reduced TGF-β1 result in delay of healing process compared with normal skin healing in DFU. Furthermore, they studied debridement intervention in DFU, proposed three types of therapeutic approaches for DFU, and demonstrated, using an agent-based model, that those types of therapeutic approaches could statistically suppress significant tissue damage in DFU. In 2008, Li et al., proposed an agent-based model for simulating inflammation of acute vocal fold injury [16]. The agent-based model quantitatively reproduced and predicted trajectories of inflammatory cytokines such as TNF-α, IL-1β and IL-10 under four-hour specific treatments, including spontaneous speech, voice rest, and resonant voice in acute vocal fold injury. Simulation results have shown theoretical individual-specific trajectories of mediator levels across treatments while revealing potential application of agent-based modeling used to design patient-specific therapies in acute vocal fold injury or expansion to other clinical domains. Also in 2008, Dancik et al., proposed an agent-based model to describe natural dynamics of immune response to L. major infection [17]. They simulated infection of macrophage by L. major infection as well as the recruitment of T cells in adaptive immunity response in the presence of chemokines, such as IL-8 to delineate underlying cellular mechanisms of L. major infection. By conducting sensitivity analysis, results indicated that strength and timing of adaptive immune response, resting macrophage speed, and transfer threshold of macrophages impact parasite load at the peak of infection. In 2011, an agent-based mode of activation of Pseudomonas aeruginosa virulence in the stressed gut was developed to characterize and translate information of the host response to microbe into a behavioral rule of computational agents [18]. Aggregated behavioral rules of computational agents, integrated by modular submodels, described intracellular pathways and cross-cells pathways in gut immunity. Model shows effects of initial Pseudomonas population on simulated host injury and measures effects of initial Pseudomonas population on gut flora and barrier function. Furthermore, the agent-based model is used to investigate the host-pathogen system as it responds to different experimental conditions which are not developed yet, such as transient intestinal ischemia, host stress, and phosphate depletion. Finally, the authors discussed the discrepancy between observed results in agent-based models and experimental results from animal models and illustrated hypotheses concerning the source of discrepancy. Another agent-based model concerning gut immunity was proposed by Mei et al., in 2012 [19]. They simulated the dynamics of gut immunity by delineating interactions among seven types of cells: epithelial cells, macrophages, dendritic cells, neutrophils, B cells, T cells and bacteria. Cell states are represented by a variable list and variable values are changed once cell states have changed. The author assigned three basic rules for changes in the states of cells: interaction with another cell, change in neighboring environment, and presence at the current state for a certain amount of time. Simulated results have shown that chemotactic movement and cytokine-induced cell-state change play critical roles in host-pathogen immune responses of gut immunity.
5. Implementing Software Platforms for Agent-Based Model
During the last decade, agent-based models are primarily used for modeling different aspects of real-world problems, such as economics, social networks, and host-pathogen interactions. Agent-based models in relation to economic and social networks deal with interactions among people and the impact of people’s aggregated behaviors on complex economic or social situation [20-22]. Agent-based models in relation to host-pathogen system, otherwise, deal with interactions among cells, associated cytokines and their impact on immune system [16-19]. The agent-based community has developed several agent-based toolkits, including packaged software and open source platforms to help researchers build their own agent-based model applications. In this section, we review the most commonly used agent-based toolkits and describe their applications.
In 2001, An used Starlogo to build an agent-based application to simulate AIR progression [11]. Starlogo and later versions such as MacStarLogo, OpenStarLogo, and StarLogo TNG, are categorized by logo family and developed from the logo programming language. Starlogo is recognized as an educational kit for building agentbased applications. This platform emulates a parallel-processing computer, allowing for simultaneous execution of multiple independent computer programs with a high-level programming language called Starlogo. Starlogo uses a natural programming language to describe real systems, thereby, making it understandable and easy to implement without extensive programming efforts. The main user interface of Starlogo is comprised of twodimensional grids. The agents can be divided into two categories: “patches” and “turtles.” “Patches” are fixed agents placed on background grids in the model workspace. “Turtles” are mobile agents that occupy a position or move freely on the surface of patches while executing certain functions or actions. Moreover, Starlogo offers a way to define agent set as “breed,” meaning that agent types with similar behaviors or under the control of the same mechanisms. “Breed” allows the modeler to define a class of agents with a set of common state variables and establish various functions or actions (autonomous behaviors) for agent types. Also, the modeler can generate output of a simulation and set parameters in a separate area from the Starlogo interface.
Researchers most frequently recommend and utilize the agent-based toolkit Netlogo [23]. Compared with other toolkits within the logo family, Netlogo has a similar programming environment, including platform interface, agent types, and programming statements. Starlogo could easily be converted to Netlogo by adjusting certain statements. Netlogo is recognized by developers of agent-based modeling as an advanced version of Starlogo because it incorporates more agent types such as “link” and, consequently could construct more sophisticated systems [24]. Furthermore, Netlogo includes a wide range of library models which could help new researchers prototype their own models. Therefore, Netlogo is recognized as the most professional platform for simulating real systems by providing a simple high-level programming language, built-in graphic interfaces, and comprehensive documentation [24].
Besides built-in, high-level programming language, agent-based platforms such as Starlogo or Netlogo, Objective-C Swarm, and its derived, Java Swarm, provide well-experienced programmers conceptual frameworks for building agent-based models. The primary advantage of Objective-C Swarm and Java Swarm is they help organize different levels of agent-based models into hierarchy and eventually integrate those small agent-based models into a complex agent-based model. This advantage plays an essential role in developing agent-based models for the immune system at the whole body level by integrating several small agent-based models at organ level. However, the disadvantages of Swam platform include lack of novice-friendly development tools, difficulty in building models because of low-level programming language, and low availability of documentation and tutorial material [24].
Recently, another popular implementing software platform for agent-based models, called Repast Symphony, is used in building agent-based simulation [25]. Mei’s research group proposed an agent-based simulator called Enteric Immunity Simulator (ENISI) Visual for modeling gut immunity [19]. ENISI Visual has an interface comprised of a series of grids. Each grid has a value indicating concentration of agents, and grid background colors change as states change. The implementer could run the simulator by setting up the initial numbers of indicators, simulation speed, steps, runs, and agent movements (random or chemotactic). ENISI Visual simulator is implemented by Java language based on Repast Symphony, a popular platform for agent-based modeling [25]. The homepage of Repast Symphony states that “Repast Symphony is an integrated, richly interactive, cross platform Java-based modeling system that runs under Microsoft Windows, Apple Mac OS X, and Linux. It supports the development of extremely flexible models of interacting agents for use on workstations and small computing clusters.” Specially, Repast Symphony could integrate the Netlogo model into Relogo. The interface of Repast Symphony is comprised of three main parts: the top line is the control panel, the left side is the user panel for setting initial values, and the right side is a visual window for observing agent movements and interactions. The advantage of Repast Symphony is that this software platform could highly customize agent behaviors and interaction among agents by incorporating a programming language such as Java. Few of current agentbased models have been built using Repast, but Repast could be a potential powerful agent-based took it with its development in functions.
Mason is recognized as a smaller and faster alternative to Repast, recently designed as a Java-based platform with a multi-agent simulation environment [24]. Compared to other agent-based platforms, Mason is recognized as a less mature simulation package but having the least execution time, which is appropriate to simulate agent behavior with much iteration for experienced programmers. We have compared implementation of different agent-based platforms in the following aspects listed in Table1
Table 1 shows that different characteristics of five agent-based simulation platforms in seven aspects. Only Netlgo has built-in agent and built-in patches which could easily start with building interactions of agents and tracking agents’ movement. Other than Netlogo and Repast, Swam, Java Swam and Mason need to write in low

Table 1. Characteristics of various types of agent-based simulation platforms.
level language with careful design of programming. Especially, Swam need to have a generator to build agents and does not have toroidal interface. Five agent-based took its have a mature color function.
When one implements an agent-based model, execution speed is crucial to determine if the agent-based platform is effective. From Railsback’s implementations of 16 versions of agent-based models [24], we summarized that Mason is the fastest agent-based platform compared to nearly all versions of the agent-based model. Repast and Netlogo closely follow in results, but Swam is the slowest agent-based platform, especially when complexity increases in model structure. In conclusion, we believe that Netlogo is an appropriate toolkit for new researchers for developing agent-based applications because of its simplified programming environment, easily implemented tool sets, well-developed library model, and well-established documentation support.
6. Advantages of Agent-Based Modeling on Infectious Disease
Agent-based modeling has been employed to describe numerous processes in immunology [6]. Complex, nonlinear biological immune processes responding to infection require integrated information to represent interaction effects among various components rather than reconstruct those processes by linearly summarizing characteristics of each single component. Compared to traditional differential equation models, Bonabeau [26] claims that agent-based modeling (ABM) is a powerful simulation modeling technique for naturally describing nonlinear relationships between components in immune responses as a whole. The author explained that ABM could simulate more complicated individual behaviors in spatial and local environments and further exhibit individual learning and adaptation by modeling and simulating behavior of the system’s constituent units and their interaction. Later, Bauer et al., [6] classified multiple applications of ABM in immunology. They reviewed various ABMs relevant to host-pathogen systems and discussed contributions to understanding immunology and disease pathology. They pointed out that ABMs are closer to the description and representation of a true biological system compared to traditional modeling techniques. By suggesting directions and velocities of cell movement in simulation, ABMs could easily provide insight into spatial or localized cell interaction in host-pathogen systems while addressing limitations of traditional modeling techniques such as ordinary differential equations and partial differential equations.
A well-detailed agent-based model derived from verified research tells a story about immune system response to various insults. First, by translating basic science evidences of infectious disease into behavioral computational agents, agent-based model is intuitive and easy to understand. Secondly, agent-based model is capable of reconstructing the interactions between cells and cytokines in relation to specific disease and therefore simulating different kinds of infectious disease. Thirdly, an agent-based model can be developed further by incorporating new types of agents or the interactions of specific types of agents with other agents, leading to greater understanding of the control mechanism for cellular behavior. Moreover, ABM is built with a random event generator, and therefore, is able to simulate the stochastic nature of immune responses to infectious disease. Analysis of various consequences of disease progression for heterogeneous patients can be accomplished by getting insight into the stochastic nature of immune responses. The randomness in an agent-based model is largely embedded in the process of agent interactions, such as one agent choosing to interact with one neighboring agent rather than another. Furthermore, agents could execute certain functions in different locations when they move or interact with other types of agents. For example, in most immune responses, neutrophils execute a series of functions such as moving rolling and adhering upon gradient to endothelial cells when they are in a blood vessel. Once they enter a nearby tissue they execute different functions and interact with various types of cytokines and immune cells. In an agent-based model, behavior of agents and the interactions of one agent with another are highly randomized and spatially-dependent which could not be described by other modeling approaches. Most importantly, in most circumstances, agent-based models are employed to support the development and design of clinical trials. By incorporating single agent-treatment or multi-agent treatment, the agent-based model could demonstrate the evidences observed in experimental design using computational results. ABM could test effects of proposed treatments, prior to clinical trials, and help in designing future potential experiments especially focused on the exploration of new therapeutic approaches. Current therapeutic experiments emphasize mediator-directed treatments. The number of those experiments largely increases with the development of new knowledge of investigated mediators.
7. Limitations of Agent-Based Modeling of Infectious Disease
Agent-based models clearly have several striking advantages; however, they also have some limitations. Firstly, an agent-based model is defined as an “instructive” tool and cannot represent real immune responses in infectious disease because it fails in one-to-one mapping of components and processes to biological systems. Biological immune responses responding to infection are recognized as a series of complex processes including both intracellular transductions (process of DNA being transferred) and intercellular pathways between cells. Those biological processes will be developed with evolved understanding and continued investigation of cellular and molecular mechanisms. Proposed agent-based models are very abstract descriptions of real systems and are still under development. The challenge of constructing an agent-based model in practice is to appropriately choose the degree of abstraction and avoid unnecessary information while incorporating essential information for recognizable results.
Also, agent behavior and interactions between agents in infectious disease are based on the understanding of basic cellular and molecular mechanisms in immune responses. However, knowledge concerning some interactions may not exist or are still under exploration [27]. Furthermore, patterns of immune responses evolved from agent interactions are observed in agent-based models and, compared with results from experimental studies, serve as validation and refinement of agent-based models. However, existing experimental studies may provide contrary information to basic biological mechanisms and patterns of immune responses, which complicates building and validating an agent-based model. Various experimental conditions, data sampling methods, and experimental designs could contribute to the conflicting results as no uniform standard exists to perform those clinical trials.
Additionally, most existing agent-based models are limited in quantitative validation of simulated results from experimental designs. Instead, they generate qualitative results to represent patterns of progression of infectious disease. However, to further validate the agent-based model, quantitative measurements are necessary to match simulated results with experimental results reported. The validation process requires a large amount of experimental data in order to incorporate or translate those data into an agent-based model. During the validation process, major difficulties occur, such as when some data is unavailable or the data format is not uniform by measurements, leading to incomplete translation of biological information into qualitative simulation.
Furthermore, an agent-based model requires high-level of computational efforts to simulate the detailed interactions among classes of agents in immune responses of infectious disease. The agent-based model is designed to describe the aggregated level of components by simulating individual agent behavior and interactions, and therefore, requires extensive computational effort and the computation efficiency is quite low. If 10 types of agents are defined in an agent-based model and each type of agent initially has 100 agents. A total of 1000 agents’ behaviors need to be encoded and decoded when executing the agent-based model. In the case of overwhelming infection, the number of bacteria (one type of agent) can explode to 108 and a large amount of computational power is therefore needed to run the model.
8. Future Direction of Agent-Based Modeling of Infection
Limitations of current agent-based models provide opportunities for future research. One of the initial steps needed in future research is to refine current agent-based models by adding more sophisticated cellular and molecular pathways in immune system when the immune system is responding to various types of infectious disease. For instance, in simulating sepsis progression, current agent-based models could be enhanced by adding anti-inflammatory pathways in both innate and adaptive immunity. The agent-based model for simulating diabetic foot ulcers could be improved by incorporating collagen contraction in the wound-healing process [14]. By taking into account more molecular interactions and transductions inside cells, the agent-based models could build a bridge between intracellular mechanism and intercellular interactions. The current agent-based models prima-

Figure 3. Basic structure of agent changing.
rily focus on the interface of blood borne endothelial cells as a platform to simulate the initial start of immune responses. The long-term goal of agent-based models is to construct various structures modeling different organs, eventually simulating the physiology at the organ level, and link immune responses at the organ level to systemic responses on a whole-body scale.
With the development of current biological experiments, experimental data could be obtained using experimental designs as inputs into the agent-based model, and quantitative results could be expected in future research. In addition, the existing agent-based models use simple arithmetic rules to regulate and control interactions and movement among agents. In future research, we hope to describe aggregated behaviors of agents in immune responses using well-formed and complete mathematical expressions derived from known and hypothesized kinetics of components of biologic systems.
The current agent-based models require a large computational effort. For instance, Netlogo models, one of many software platforms for the agent-based model, are limited to a few thousand agents running abstract rules on a high-performance computer [28]. In particular, a large number of repetitive local interactions among agents greatly increase the running time of the agent-based model. To reduce this computational hurdle, one could use agent-owned variables (local variables) to define each type of agent [13]. Values of agent-owned variables will be updated every time period predefined in the model. Similarly, the number of agent types will be updated corresponding to the change in the agent-owned variables. The relationship between the change in number of cells and change in the agent-owned variable is described in Figure 3. Figure 3 shows that agent-specific variables induce the change in the number of agents.
Similarly, dynamic agent compression allows a set of homogeneous agents stored in compact bins to make the model more efficient in its use of memory and computational cycles, therefore allowing the user to scale up complexity of the model and run the model more efficiently by increasing execution speeds [29]. Furthermore, the Gillespie algorithm proposed the generation of a statistically correct trajectory for stochastic simulation and streamlining the execution of computational steps [30]. More computational algorithms, as well as enhancement of computer power, are expected to implement multi-scale agent-based models. Parallel computers also have a potential to improve ABM applications in the future.
Acknowledgements
I would like to thank Dr. Stephen Chapes from Department of Biology at Kansas State University and Dr. Steven Simpson from University of Kansas Medical Center for helping in developing the biological models.
[2] Wikipedia.org. http://en.wikipedia.org/wiki/Infectious_disease
[3] R. Kumar, G. Clermont, Y. Vodovotz and C. C. Chow, “The Dynamics of Acute Inflammation,” Journal of Theoretical Biology, Vol. 230, No. 2, 2004, pp. 145-155. http://dx.doi.org/10.1016/j.jtbi.2004.04.044
[4] A. Reynolds, J. Rubin, G. Clermont, J. Day, Y. Vodovotz and G. B. Ermentrout, “A Reduced Mathematical Model of the Acute Inflammatory Response: I. Derivation of Model and Analysis of Anti-Inflammation,” Journal of Theoretical Biology, Vol. 242, No. 1, 2006, pp. 220-236. http://dx.doi.org/10.1016/j.jtbi.2006.02.016
[5] R. Axelrod, “The Complexity of Cooperation: Agent-Based Models of Competition and Collaboration,” 1th Edition, Princeton University Press, 1997.
[6] J. M. Epstein and R. L. Axtell, “Growing Artificial Societies: Social Science from the Bottom Up,” 1th Edition, MIT Press, 1996.
[7] A. L. Bauer, C. A. A. Beauchemin and A. S. Perelson, “Agent-Based Modeling of Host-Pathogen Systems: The Successes and Challenges,” Information Sciences, Vol. 179, No. 10, 2009, pp. 1379-1389. http://dx.doi.org/10.1016/j.ins.2008.11.012
[8] T. K. Kar, “Stability Analysis of a Prey-Predator Model Incorporating a Prey Refuge,” Communications in Nonlinear Science and Numerical Simulation, Vol. 10, No. 6, 2005, pp. 681-691. http://dx.doi.org/10.1016/j.cnsns.2003.08.006
[9] R. C. Bone, R. A. Balk, F. B. Cerra, R. P. Dellinger, A. M. Fein and W. A. Knaus, “Definitions for Sepsis and Organ Failure and Guidelines for the Use of Innovative Therapies in Sepsis,” Chest, Vol. 101, No. 6, 1992, pp. 1644-1655.
[10] M. P. Glauser, “Pathophysiologic Basis of Sepsis: Considerations for Future Strategies of Intervention,” Critical Care Medicine, Vol. 28, No. 9, 2000, pp. 84-88.
[11] American College of Chest Physicians/Society of Critical Medicine Consensus Committee, “Definitions for Sepsis and Organ Failure and Guidelines for the Use of Innovative Therapies in Sepsis,” Critical Care Medicine, Vol. 20, No. 6, 1992, pp. 864-874. http://dx.doi.org/10.1097/00003246-199206000-00025
[12] G. An, “Agent-Based Computer Simulation and SIRS: Building a Bridge between Basic Science and Clinical Trials,” Shock, Vol. 16, No. 4, 2001, pp. 266-273. http://dx.doi.org/10.1097/00024382-200116040-00006
[13] G. An, “In Silico Experiments of Existing and Hypothetical Cytokine-Directed Clinical Trials Using Agent-Based Modeling,” Critical Care Medicine, Vol. 32, No. 10, 2004, pp. 2050-2060. http://dx.doi.org/10.1097/01.CCM.0000139707.13729.7D
[14] J. Wu, D. Ben-Arieh and Z. Z. Shi, “An Autonomous Multi-Agent Simulation Model for Acute Inflammatory Response,” International Journal of Artificial Life Research, Vol. 2, No. 2, 2011, pp. 105-121. http://dx.doi.org/10.4018/jalr.2011040106
[15] X. Dong, P. T. Foteinou, S. E. Calvano, S. F. Lowry and I. P. Androulakis, “Agent-Based Modeling of Endotoxin-Induced Acute Inflammatory Response in Human Blood Leukocytes,” PLoS One, Vol. 5, No. 2, 2011, Article ID: e9249. http://dx.doi.org/10.1371/journal.pone.0009249
[16] Q. Mi, B. Rivière, G. Clermont, D. L. Steed and Y. Vodovotz, “Agent-Based Model of Inflammation and Wound Healing: Insights into Diabetic Foot Ulcer Pathology and the Role of Transforming Growth Factor-β1,” Wound Repair and Regeneration, Vol. 15, No. 5, 2007, pp. 671-682. http://dx.doi.org/10.1111/j.1524-475X.2007.00271.x
[17] N. Y. K. Li, K. Verdolini, G. Clermont, Q. Mi, E. N. Rubinstein, P. A. Hebda and Y. Vodovotz, “A Patient-Specific in Silico Model of Inflammation and Healing Tested in Acute Vocal Fold Injury,” PLoS One, Vol. 3, No. 7, 2008, Article ID: e2789. http://dx.doi.org/10.1371/journal.pone.0002789
[18] G. M. Dancik, D. E. Jones and K. S. Dorman, “An Agent-Based Model for Leishmania Major Infection,” Unifying Themes in Complex System, pp. 243-250.
[19] J. B. Seal, J. C. Alverdy, O. Zaborina and G. An, “Agent-Based Dynamic Knowledge Representation of Pseudomonas aeruginosa Virulence Activation in the Stressed Gut: Towards Characterizing Host-Pathogen Interactions in Gut-Derived Sepsis,” Theoretical Biology and Medical Modelling, Vol. 8, No. 33, 2011, pp. 1-34.
[20] Y. G. Mei, R. Hontecillas, X. Y. Zhang, K. Bisset, S. Eubank, S. Hoops, M. Marathe and J. Bassaganya-Riera, “ENISI Visual, an Agent-Based Simulator for Modeling Gut Immunity,” IEEE International Conference on Bioinformatics and Biomedicine, Philadelphia, 4-7 October 2012, pp. 1-5.
[21] T. Lux and M. Marchesi, “Volatility Clustering in Financial Market: A Micro-Simulation of Interacting Agents,” International Journal of Theoretical and Applied Finance, Vol. 3, No. 4, 2000, pp. 675-702. http://dx.doi.org/10.1142/S0219024900000826
[22] B. LeBaron, “Agent-Based Computational Finance: Suggested Readings and Early Research,” Journal of Economic Dynamics and Control, Vol. 24, No. 5-7, 2000, pp. 679-702. http://dx.doi.org/10.1016/S0165-1889(99)00022-6
[23] A. M. EI-Sayed, P. Scarborough, L. Seemann and S. Galea, “Social Network Analysis and Agent-Based Modeling in Social Epidemiology,” Epidemiologic Perspectives and Innovations, Vol. 9, No. 9, 2012, pp. 1-9.
[24] Netlogo, Software Version 4.0.4. http://ccl.northwestern.edu/netlogo
[25] S. F. Railsback, “Agent-Based Simulation Platforms: Review and Development Recommendations,” Simulation, Vol. 82, No. 9, 2006, pp. 609-623. http://dx.doi.org/10.1177/0037549706073695
[26] Repast. http://repast.sourceforge.net/
[27] E. Bonabeau, “Agent-Based Modeling: Methods and Techniques for Simulating Human Systems,” PNAS, Vol. 99, No. 3, 2002, pp. 7280-7287. http://dx.doi.org/10.1073/pnas.082080899
[28] M. Aziz, A. Jacob, W. L. Yang, A. Matsuda and P. Wang, “Current Trends in Inflammatory and Immunomodulatory Mediators in Sepsis,” Journal of Leukocyte Biology, Vol. 93, No. 3, 2013, pp. 1-14. http://dx.doi.org/10.1189/jlb.0912437
[29] G. An, “Introduction of an Agent-Based Multi-Scale Modular Architecture for Dynamic Knowledge Representation of Acute Inflammation,” Theoretical Biology and Medical Modeling, Vol. 5, 2008, p. 11. http://dx.doi.org/10.1186/1742-4682-5-11
[30] S. Wendel and C. Dibble, “Dynamic Agent Compression,” Journal of Artificial Societies and Social Simulation, Vol. 10, No. 2, 2007, pp. 1-16.
[31] L. Harris and P. Clancy, “A Partitioned Leaping Approach to Multi-Scale Modeling of Chemical Reaction Dynamics,” Journal of Chemical Physics, Vol. 125, No. 14, 2006, pp. 1-10. http://dx.doi.org/10.1063/1.2354085
NOTES
*Corresponding author.