Isolation and Characterization of Hydrocarbon Utilizing Yeast (HUY) Isolates from Palm Wine ()
Received 27 December 2015; accepted 28 February 2016; published 2 March 2016

1. Introduction
Diverse microbial population bacteria, yeasts or molds have been reported for degradation of hydrocarbons. The reported efficiency of biodegradation ranged from 6% to 82% for fungi, 0.13% to 50% for soil bacteria, and 0.003% to 100% for marine bacteria [1] .
Yeasts are eukaryotic microorganisms, classified in the kingdom fungi, with about 1500 species currently identified and described. They are estimated to be 1% of all fungal species [2] and measures up to 3 - 4 µm in diameter some can reach over 40 µm depending on the species [3] .
Most young yeast colonies are moist, somewhat slimy in appearance and may also appear mealy. The colour of most colonies are whitish, cream-coloured or pink and change little with age, but others become dry and wrinkled. Yeasts are oxidative, fermentative or both. Oxidative yeast (film yeasts) may grow as a film or scum on the surface of a liquid medium where as fermentative yeasts grow throughout the liquid [4] . They are chemoorganotrophs, hence they use organic compounds as source of energy [5] , they could be aerobes, facultative anaerobes but never strict or obligate anaerobes [6] [7] .
Yeasts are widely dispersed in the environment. They grow best in neutral or slightly acidic pH environment. Some are normal flora on skin surfaces; others are parasitic or symbiotic and are mostly common in environments where there is a sugar-rich material. Some are associated with soil and insects [7] [8] .
[9] reported that palm wine is a suspension of different types of microorganisms including bacteria, filamentous fungi, and yeasts. Yeasts occur in palm wine as indigenous micro flora and are mainly from the genus Saccharomyces, Schizosaccharomyces, Pichia, Candida, Kleockera, Hansenula, Endomycopsis and Saccharomycoides [10] - [12] .
Several studies have reported on the capabilities of different yeasts to utilize hydrocarbons. The yeasts implicated in previous studies belong to the genera: Candida, Clavispora, Pichia, Sporobolomyces, Sporidiobolus, Stephanoascus, Debaryomyces, Lodderromyces, Leucosporidium, Metschnikowia, Rhodotorula, Rhodosporidium, Trichosporon and Yarrowia [7] [13] [14] . Irrespective of the reports from these studies, there is a dearth of information on the capability of yeast obtained from non oil polluted environment to degrade hydrocarbons which underscores the relevance of this study.
The study aimed at isolation and identification of yeast species in palm sap (palm wine), to ascertain their ability to degrade petroleum hydrocarbons. The objectives of this study were to:
1) Isolate and characterize yeasts from palm wine using morphological and molecular method.
2) Test the isolates for biodegradation of petroleum hydrocarbons.
2. Materials and Methods
2.1. Sample Collection
The crude oil used in this research was the Nigerian Bonny light crude oil obtained from Shell Petroleum Development Company (SPDC) Limited, Port Harcourt, Nigeria.
Yeasts used in this study were isolated from fresh palm wine gotten from raffia palm (Raphiaraphia) by palm wine tappers in Choba, Obia/Akpo Local Government Area, Rivers State. The samples were transported immediately to the laboratory in sterile containers packed in coolers with ice packs for analysis.
2.2. Experimental Design
2.2.1. Isolation of Test Organisms
One milliliter of palm wine was inoculated into 200 ml sterilized mineral salts broth [15] as described by [16] . Crude oil samples were filtered and autoclaved at 121˚C for 15 mins for sterility and when cooled added to the inoculums and swirled for proper mixing at a concentration of 1% (v/v). Incubation was done for fourteen days at room temperature (25˚C ± 2˚C) without shaking.
The spread plate technique was used to inoculate the viable culturable isolates. A glass spreading rod that has been sterilized in alcohol and flamed was used for this procedure. A 0.1 ml of the above enrichment medium was inoculated onto mineral salt agar plates in triplicates.
According to [17] , sterile filter papers (Whatman No. 1) saturated with crude oil was placed on the inside of the cover plate of each Petri dish. The inoculated Petri dishes were kept in an inverted position and incubated at 25˚C ± 2˚C for five days. These filter papers supplied the hydrocarbons by the vapour phase transfer to the sub- cultured isolates.
The unadapted microorganisms were obtained by inoculating 0.1 ml of the fresh palm wine onto sterile mineral salt agar plates in triplicates using the spread plate method. Agar plates contained crude oil-soaked filter paper and incubated at 25˚C ± 2˚C for fourteen days in inverted position.
Viable isolates from the above media were purified using Sabouraud dextrose agar (balanced peptone water No.1 10 g, Dextrose 40 g, agar No.2 12 g, pH 5.6 ± 0.2). The adapted and unadapted pure isolates obtained were aseptically inoculated into the agar slants in the McCartney bottles. These were incubated at 25˚C ± 2˚C for 48 hours. These stock cultures were preserved in the refrigerator at 4˚C for future use. Yeast isolates obtained were prefixed with the letters Pw (Palm wine) e.g. Pw 01, Pw 02 for the first and second isolates respectively. Identification of yeast isolates were based on macroscopic and microscopic examination of the morphological and biochemical properties [18] . Wet mounts of a 48 hour culture were used for microscopy.
Sugar fermentation test was carried out to determine the ability of the isolates to metabolise some carbohydrates such as glucose, lactose, galactose, trehalose, inositol, maltose, dulcitol, sucrose and raffinose with the production of either acid, gas or both.
2.2.2. Screening for Biodegradation Potentials
The modified method of [19] , was used for the screening test. A loopful of a 48 hour culture of the isolates were inoculated into a sterile 100 ml of Bacto Bushnell Haas broth containing 1 ml of sterile crude oil (1% v/v) and 1 ml of redox indicator (2% v/v of 2,6-dichlorophenol indophenols). A control was also set up containing no microorganisms. The set up was left for 15 days. Total hydrocarbon content (Oil and Grease) was determined according to API-RP45 method using a Spectrophotometer. The sample was extracted twice with 1:10 ratio of Xylene to sample. The combined extract after centrifuging was read in the spectrophotometer using Xylene as the reference material. The spectrophotometer had been previously calibrated with crude oil. Readings obtained from the spectrophotometer were traced out on the calibration graph and used to calculate the concentration of THC (Oil and Grease) in mg/l [20] . Total Hydrocarbon Content was absorbed at 420 nm using the spectrophotometer Corning 253.
Calculation:
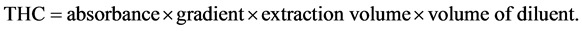
2.2.3. Molecular Identification of Test Isolates
Pure cultures of the potential strains maintained on Sabouraud dextrose agar slants in McCartney bottles were identified at the Biotechnology Centre, Federal University of Agriculture, Abeokuta, Ogun state (FUNAAB) and Microgen U.S.A. employing deoxyribonucleic acid (DNA) extraction, deoxyribonucleic acid (DNA) sequencing and sequence blasting on National Centre for Biotechnology Information (NCBI). The DNA extraction was carried out using the NORGEN BIOTEK CORP, Fungi/Yeast Genomic DNA Isolation Kit.
One milliliter of the washed microorganisms was transferred to a microcentrifuge tube and centrifuged using spectrafuge 24D, Labnet international, Inc. at 14,000 rpm gentle vortexing using the Stuart vortex mixer, SA8. Agarose gel electrophoresis was used to determine the quality and integrity of the DNA by size fractionation on 1.0% agarose gels. Agarose gels (biotechnology grade) were prepared by dissolving and boiling 1.0 g agarose in 100 ml 0.5× tris acetate ethylene diamine tetraacetic acid (TAE) buffer solution. The gels were allowed to cool down to about 45˚C and 10 µL of 5 mg/ml ethidium bromide was added, mixed together before pouring it into an electrophoresis chamber set with the combs inserted. After the gel has solidified, 3 µL of the DNA with 5 µL sterile distilled water and 2 µL of 6× loading dye was mixed together and loaded in the well created. Electrophoresis was done using the Consort EV231 electrophoresis machine at 80 V for 2 hours. The integrity of the DNA was visualized and photographed on UV light source by the UV documentation.
Polymerase chain reaction (PCR) was carried out using the MJ research thermal cycler (PTC-200). The amplicon was further purified prior to sequencing using 2 M sodium acetate wash technique.
The primer used for the reaction is 18S forward and reverse. The samples were loaded on the machine and the data in form A, C, T, and G were released.
The sequencing was carried out using the applied biosystem; ABI 3130X1 model. The isolates’ genes were sequenced using the ITS 1 and ITS 2 primer [21] . The primer sequences are as shown below:
Primer Name Sequence (5′-3′).
ITS 1 TCCGTAGGTGAACCTGCGG
ITS 2 GCTGCGTTCTTCATCGATGC
Sequence results obtained from above were compared with known sequences in the Genbank using the Basic Local Alignment Search Tool (BLAST) of the National Centre for Biotechnology Information (NCBI). Species were identified based on the percentage (%) similarity with the known species (sequences in the database).
3. Results and Discussion
In this study, thirteen yeast isolates were isolated from the enriched medium and fresh palm wine. Results from the enrichment medium (adapted microorganisms) and the palm wine (unadapted microorganisms) showed that microorganisms were of the same type based on colonial and cellular morphologies (Table 1). According to the colonial morphology on solid media, it was found that both isolates (Pw 1 and Pw 3) have cream colour but Pw 3 appears dull with age. They (adapted and unadapted microorganisms) also exhibited same reactions to biochemical test (Table 2). The yeast isolates were Gram positive, metabolized glucose with the production of acid and gas. Yeast isolates were identified tentatively as Saccharomyces species (Pw 1) and Schizosaccharomy- ces species (Pw 3) (Plate 1). The yeasts morphology under Celestron digital microscope was studied and found that Schizosaccharomyces species cells were small and Saccharomyces species cells were larger and spherical in shape.
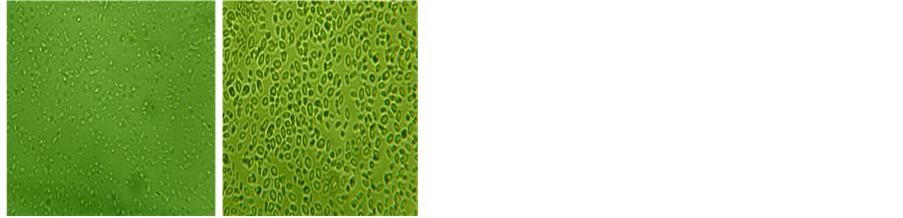
(a) (b)
Plate 1. Celestron digital microscope micrographs of yeast cells at 25˚C ± 2˚C. (a) Schizosaccharomyces species; (b) Saccharomyces species.
![]()
Table 1. Cultural and morphological characteristics of yeast isolates.
KEY: A/G: Acid and gas production; A/−: Acid and no gas production; +: Positive; −: Negative.
3.1. Screening for Biodegradation Potentials
All yeast isolates were subjected to screening for biodegradation potentials using the 2, 6-dichlorophenol indophenols as indicator agent. There was a gradual colour change in the setup from deep blue (the colour of the indicator agent) to purple to maroon and finally colourless. This colour change suggests that yeast isolates have biodegradative potentials. During the screening for biodegradation potentials, the total hydrocarbon content (THC) was reduced suggesting that yeast isolates were potential hydrocarbon degraders. Previous studies have shown that the bio-augmentation technology could significantly improve the efficiency of bioremediation (Pozdnyakova et al., 2008; Jacques et al., 2008). Irrespective of the findings, other researchers have reported on the ineffectiveness of exogenous microorganisms to repair the oil contaminated soil (Marianoand Kataoka, 2007; Ueno et al., 2006; Liu and Wang, 2008). In this present study, the addition of exogenous yeasts isolated from palm wine biodegraded petroleum hydrocarbon thereby corroborating earlier reports in this regard. This research was based on the principle of allochthonous bio-augmentation since the yeasts were isolated from a different source other than petroleum hydrocarbon contaminated sites.
The capability of several yeast species to use n-alkanes and other aliphatic hydrocarbons as a sole source of carbon and energy is reportedly mediated by the existence of multiple microsomal Cytochrome P450 forms. These cytochrome P450 enzymes had been isolated from yeast species such as Candida maltosa, Candida tropicalis, and Candida apicola (Scheuer et al., 1998). Sood and Lal (2009) reported that the ability of the strain to grow in MSM with hydrocarbons as the sole carbon source is indicative of its ability to utilize the hydrocarbons.
3.2. Molecular Identification of Yeast Isolates
Yeast isolates’ DNA were isolated with the DNA extraction kit, amplified using the polymerase chain reaction and purified. Using the ITS 1 and ITS 2 primers, the genes were sequenced and BLAST carried out to compare these with those in GenBank as described by Mrinalini and Jayanthi (2011). The yeast isolates that were identified tentatively as Schizosaccharomyces species and Saccharomyces species when molecularly identified based on the 18S-rRNA were Candida adriatica ZIM 2468 and Candida taoyuanica MYA-4700 as shown in Table 3.
The study showed that isolates from the enrichment medium (adapted microorganisms) and the palm wine (unadapted microorganisms) were of the same type based on colonial and cellular morphologies (Table 1). Yeast isolates were identified tentatively and with the Celestron digital microscope as Saccharomyces species and Schizosaccharomyces species.
![]()
Table 3. Molecular identification by 18S-rRNA.
Molecular identification was carried out by sequencing the isolates’ genes using the 18S-rRNA for fungi [21] . It will interest us to know that when the sequences were marched with the GenBank database using the Basic Local Allignment Search Tool (BLAST) of the National Centre for Biotechnology Information (NCBI), Schizosaccharomyces species showed 98% similarity with Candida adriatica ZIM 2468 and Saccharomyces species 100% with Candida taoyuanica MYA-4700. In the works of Tuntiwongwanich and Leenanon (2009), Candida species were also isolated from palm wine obtained from different palms.
Ability of the isolates to produce colour change in the medium during the screening for biodegradative potential is due to the reduction of the indicator agent; 2,6-dichlorophenol indophenols by the oxidized products of hydrocarbon degradation. The colour change supports the fact that the isolates are potential hydrocarbon utilizers. [19] , in a related work also, observed a change in colour from deep blue to colourless. [22] , said that another criterion to determine isolates’ biodegradative potentials is the rupture of oily surface of the culture medium which we also, observed during the study. The reduction in the residual total hydrocarbon content (THC) during the screening suggests that isolates are hydrocarbon utilizers.
It is interesting to note that yeast isolates normally present in palm wine; a non-oil impacted environment, has the capability to utilize crude oil/petroleum hydrocarbons as sole source of carbon and energy. [17] , in their work isolated yeasts capable of utilising kerosine and diesel as sole source of carbon from palm wine.
According to [11] , microbial communities exposed to hydrocarbons adapt to this exposure through selective enrichment and genetic changes resulting in an increase in hydrocarbon-degradation. This pre-exposure of microorganisms make them better suited to degrade the pollutant through higher growth and reproduction and more efficient metabolism thus maximizing the rate of hydrocarbon removal from the soil.
4. Conclusions
Thirteen yeast isolates were collected from the enrichment medium and palm wine in this study. Two different strains were identified as Candida adriatica ZIM 2468 and Candida taoyuanica MYA 4700 based on the use of 18S rRNA for fungi. Under the Celestron digital microscope, Candida adriatica cells were small, oval creamy but turn gold with age while Candida taoyuanica cells were larger and ovoid. Both strains were hydrocarbon utilizers as observed in the biodegradation screening which were indicated by the change in colour from deep blue to colourless and rupturing of the oily surface of the culture medium.
The exogenous yeast isolated from palm wine that is not polluted with petroleum hydrocarbons was screened for biodegradation potentials; the total hydrocarbon content (THC) was reduced suggesting that yeast isolates were potential hydrocarbon degraders.
Authors’ Contributions
This work was carried out in collaboration between all authors. Authors T. L. Ataikiru and P. O. Okerentugba designed the study, performed the statistical analysis, wrote the protocol, and wrote the first draft of the manuscript. Author P. O. Okerentugba is the supervisor under whose guide the entire execution of the work and corrections were done to this present standard. Author T. Ichor effected the corrections of the study, managed the literature searches, was responsible for sending the article and effected all the corrections that arose thereof from the publishers. The authors declare no competing interest.