Folding and Unfolding Simulations of a Three-Stranded Beta-Sheet Protein ()

1. Introduction
Protein is a polypeptide in a living organism [1]. It is a linear polymer, built of twenty different amino acids (or residues), with a defined amino-acid sequence. Each amino acid consists of the intrinsic side-chain and the common backbone. Because different amino acids have a common backbone structure, their side-chains determine their nature―hydrophilic or hydrophobic. Hydrophobic amino acids of an unfolded protein in water drive it to fold into its three-dimensional native structure, determining its biological function. Two amino acids, glycine (G, Gly) and proline (P, Pro), are neutral. Usually, alanine (A, Ala), valine (V, Val), leucine (L, Leu), isoleucine (I, Ile), cysteine (C, Cys), methionine (M, Met), phenylalanine (F, Phe), tyrosine(Y, Tyr), and tryptophan (W, Trp) are classified as hydrophobic amino acids. Similarly, serine (S, Ser), threonine (T, Thr), asparagine (N, Asn), glutamine (Q, Gln), aspartic acid (D, Asp), glutamic acid (E, Glu), lysine (K, Lys), arginine (R, Arg), and histidine (H, His) are considered as hydrophilic amino acids. Among them, aspartic acid and glutamic acids are negatively charged, whereas lysine, arginine, and histidine are positively charged.
The primary structure of a protein is the one-dimensional amino-acid sequence of the protein, which is translated from the nucleic-acid sequence of the corresponding gene. For example, the primary structure of the brain neuropeptide Met-enkephalin, composed of five amino acids, is Tyr-Gly-Gly-Phe-Met [2]. The usual local conformations of backbones, such as alpha-helix and beta-sheet, of proteins correspond to the secondary structures of proteins. Alpha-helix, the most abundant secondary structure, is easily formed between amino acids neighboring in the sequence of a protein via hydrogen bonds between the backbone pairs, carboxyl oxygen and nitrogen with hydrogen. The right-handed alpha-helix has 3.6 residues per turn with the length 5.4 Å. The formation process of alpha-helix is well understood [3]. Each kind of protein has its unique three-dimensional folded structure, called the tertiary (native) structure. Information on the tertiary native structure of a protein is quite crucial in understanding its biological function and role.
Understanding the folding processes of a protein into its tertiary native structure only with its primary structure (amino-acid sequence information) is a long-standing challenge in modern science [4]. Understanding these folding processes is particularly important in this post-genomic era. Protein folding processes play the most important role in controlling a wide range of cellular functions. The failure of a proper protein folding results in the malfunction of biological systems, leading to various diseases. Although extensive experimental and computational studies on protein folding processes have been performed, many aspects of the processes are poorly understood [4].
Computer simulations have been carried out to study protein folding processes [5]. However, simulation of protein-folding processes with an atomistic model is a very difficult task. Usually, direct folding simulations have been mainly focused on simple models, such as lattice models, models where only native interactions are included (Go-type models), and a model with discrete energy terms whose parameters are optimized separately for each protein. Alternative indirect approaches have also been proposed including unfolding simulations starting from the folded state of a protein. Because protein folding simulation requires a very long time scale, protein unfolding simulation has been one of the most popular approaches. However, it is not clear whether we understand protein folding processes from unfolding simulations perfectly.
One of the most regular conformations adopted by proteins is the beta-sheet whose basic unit is the beta- strand. It is not stable itself whereas a single alpha-helix is stable itself. Frequently, this unstability of a single beta-strand results in formation of amyloid fibrils and various fatal diseases. It is difficult to understand the formation processes and stability of proteins with beta-strands. Betanova [6] is a monomeric, beta-sheet protein consisting of three antiparallel beta-strands. Betanova has twenty amino acids, and its primary structure is given by Arg-Gly-Trp-Ser-Val-Gln-Asn-Gly-Lys-Tyr-Thr-Asn-Asn-Gly-Lys-Thr-Thr-Glu-Gly-Arg.In this article, we perform and compare both folding simulations (starting from non-native conformations) and unfolding simulations (starting from the tertiary native structure) for betanova using the united-residue force field [7] and the most popular computer simulation method, Metropolis Monte Carlo algorithm [8].
2. Computational Methods
In the united-residue force field [7], a protein chain is represented by a sequence of alpha-carbon (Cα) atoms connected by virtual bonds with attached united side-chains (SC) and united peptide group slocated in the middle of the Cα-Cα virtual bonds. All the virtual bonds are fixed in length; the Cα-Cα length is set to 3.8 Å, and the Cα-SC lengths are given for each amino-acid type. The energy function of a protein in the united-residue force field is given by
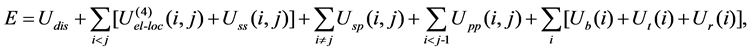 (1)
(1)
where Udis denotes the energy term to form a disulfide bridge and U(4)el-loc is the four-body interaction term. Uss(i, j) represents the mean free energy of the hydrophobic (hydrophilic)interaction between the side-chains i and j, Usp(i, j) corresponds to the excluded-volume interaction between the side-chain i and the peptide group j, and Upp(i, j) accounts for the electrostatic interaction between the peptide groups i and j. The terms Ub(i), Ut(i) and Ur(i) denote the energies of virtual angle bending, virtual dihedral angle torsions, and side-chain rotamers, respectively. The parameters of the united-residue force field were optimized simultaneously for four proteins; betanova, zink-finger based beta-beta-alpha motif 1fsd (28 amino acids, one beta-hairpin and one alpha-helix), villin headpiece protein subdomain HP-36 (36 amino acids, three alpha-helix bundle), and fragment B of staphylococcal protein A (46 amino acids, three alpha-helix bundle). The parameters were adjusted in such a way that the native-like conformations are more favored than the others energetically. In [7], the procedures to obtain the optimized parameters used in this article are described in detail.
The perturbed conformation is accepted according to the change in the energy, following Metropolis Monte Carlo algorithm [8]. For betanova, 100 independent unfolding simulations (starting from the tertiarynative structure) with 105 Monte Carlo steps (shortly, MCS) were run at a fixed temperature. We divided 105 MCS into 28 intervals (first ten 102 MCS, subsequent nine 103 MCS, and the next nine 104 MCS), and took average in each interval. These averages were again averaged over 100 independent unfolding simulations at a given temperature. Also, 200 independent folding simulations (106 MCS for each run) were performed at a fixed temperature for betanova [9]. For folding simulations starting from non-native conformations, we divided 106 MCS into 28 intervals (first ten 103 MCS, subsequent nine 104 MCS, and the next nine 105 MCS), and the averages are taken over the whole conformations for each interval. These averages are averaged again over 100 independent computer simulations starting from random conformations. The same procedure is repeated for 100 independent computer simulations starting from fully extended conformations.
3. Computational Results
During all Monte Carlo simulations the values of the root-mean-square deviation (RMSD) from the experimental structure and the radius of gyration Rg were calculated using Cα coordinates. The fraction of the native contacts ρ is also measured during all computer simulations [10]. The values of ρ = 1 and ρ = 0 mean the experimental structure and a completely disordered conformation, respectively. RMSD, the radius of gyration, and the fraction of native contacts are the most popular reaction coordinates in understanding the folding and unfolding processes between the primary structure (one-dimensional amino-acid sequence) and the tertiary (three-dimen- sional) native structure.
Figure 1 shows two different folding pathways at T = 40 (arbitrary units) and the unfolding pathways at two different temperatures T = 100 and T = 200, between the folded native structure and unfolded conformations, in the reaction coordinates ρ and Rg (in units of Å) for betanova. The folding pathways are obtained from 200 independent computer simulations at T = 40. The green triangles represent the averages of 100 folding pathways starting from random conformations. The red inverted triangles are the averages over 100 folding pathways starting from fully extended conformations (with ρ = 0, Rg = 19.9 Å, and RMSD = 16.4 Å). The unfolding pathways are obtained from 100 independent computer simulations, starting from the folded native structure (with ρ = 0.97, Rg = 7.6Å, and RMSD = 1.6 Å), at a fixed temperature. The black circles represent the averages of 100 unfolding pathways at T = 100, and the blue squares are the averages over 100 unfolding pathways at T = 200. As shown in Figure 1, the unfolding pathways are similar even for quite different temperatures T = 100 and T = 200. The unfolding pathways are almost identical from the point of ρ = 0.97 and Rg = 7.6 Åto the point of ρ = 0.75 and Rg = 8 Å, corresponding to a native-like conformation of betanova, still maintaining the antiparallel three-stranded beta-sheet. Two different folding pathways converge at the point of ρ = 0.3 and Rg = 9.5 Å, corresponding to the collapsed unfolded conformations. From this point, they are almost identical. Finally, the folding pathways and the unfolding pathways meet at the point of ρ = 0.75 and Rg = 8 Å. Between ρ = 0.3 and ρ = 0.75, the unfolding pathways lie slightly above the folding pathways for the same ρ value.
Figure 2 shows the folding pathways and the unfolding pathways between the folded native structure and the unfolded conformations in the reaction coordinates, ρ and RMSD (in units of Å), for betanova. Around ρ = 0.2, four different pathways converge. Between ρ = 0.2 and ρ = 0.5, the folding pathways and the unfolding pathways are similar. For ρ > 0.55, the folding pathways lie above the unfolding pathways for the same ρ value, and the difference in RMSD values between the folding pathways and the unfolding pathways may become larger as ρ increases.
Figure 3 shows the folding pathways and the unfolding pathways between the folded native structure and the unfolded conformations in the reaction coordinates RMSD (in units of Å) and Rg (in units of Å) for betanova. Between RMSD = 7 Å and 9Å, the folding pathways starting from fully extended conformations are nearly identical to two different unfolding pathways. For RMSD < 7 Å, the folding pathways lie slightly below the unfolding pathways for the same RMSD value. Figure 3 suggests that the folding pathways meet the unfolding pathways around the point of RMSD = 1.6 Å and Rg = 7.6 Å.
![]()
Figure 1. Folding and unfolding pathways between the folded native structure and unfolded conformations in the reaction coordinates, the fraction of native contacts ρ and the radius of gyration, for betanova. The values of ρ = 1 and ρ = 0 mean the experimental structure and a completely disordered conformation, respectively. The folding pathways are obtained from 200 independent computer simulations at T = 40. The (green) triangles represent the averages of 100 folding pathways starting from random conformations. The (red) inverted triangles are the averages over 100 folding pathways starting from fully extended conformations. The unfolding pathways are obtained from 100 independent computer simulations, starting from the folded native structure, at a fixed temperature. The (black) circles represent the averages of 100 unfolding pathways at T = 100. The (blue) squares are the averages over 100 unfolding pathways at T = 200.
![]()
Figure 2. Folding and unfolding pathways in the reaction coordinates, the fraction of native contacts and the root-mean- square deviation (RMSD), for betanova. The (green) triangles, the (red) inverted triangles, the (black) circles, and the (blue) squres represent the folding pathways starting from random conformations at T = 40, the folding pathways starting from fully extended conformations at T = 40, the unfolding pathways at T = 100, and the unfolding pathways at T = 200, respectively.
![]()
Figure 3. Folding and unfolding pathways in the reaction coordinates, the root-mean-square deviation and the radius of gyration, for betanova. The (green) triangles, the (red) inverted triangles, the (black) circles, and the (blue) squares represent the folding pathways starting from random conformations at T = 40, the folding pathways starting from fully extended conformations at T = 40, the unfolding pathways at T = 100, and the unfolding pathways at T = 200, respectively.
4. Conclusion
We have performed 200 independent folding simulations (starting from non-native conformations) and 200 idependent unfolding simulations (starting from the tertiary native structure) using the united-residue force field and Metropolis Monte Carlo algorithmfor betanova (three-stranded antiparallel beta-sheet protein).From these extensive computer simulations, we have obtained two representative folding pathways and two representative unfolding pathways in the reaction coordinates such as the fraction of native contacts, the radius of gyration, and the root-mean-square deviation.The folding pathways and the unfolding pathways are similar each other. The largest deviation between the folding pathways and the unfolding pathways results fromthe root-mean-square deviation near the folded native structure. Therefore, we may conclude that unfolding computer simulations capture the essentials of folding simulations.
Acknowledgements
This was supported by Korea National University of Transportation in 2015.