1. Introduction
18F-fluorodeoxyglucose (FDG) positron emission tomography (PET) is widely used for the diagnosis and staging of malignant tumors. In addition, FDG-PET can be used to plan radiation therapy and assess treatment effectiveness. More than 1,700,000 clinical PET and positron emission tomography/computed tomography (PET/CT) studies were performed in the USA in 2011 [1] .
Visual assessment and quantitative measurements based on region of interest, such as maximum standardized uptake value (SUVmax), have been clinically used to diagnose and stage malignant tumors using FDG-PET. FDG uptake has been studied thoroughly to allow its use as an imaging biomarker in oncology. One such application is tumor segmentation on FDG-PET images to quantify cancer aggressiveness. For example, it has been reported that metabolic tumor volume and total lesion glycolysis, based on functional tumor volume assessed from FDG- PET images, were more reliable predictors of tumor prognosis than SUVmax [2] - [4] . Recent advances in radiation therapy have enabled the precise delivery of a high dose to the target tumor; FDG-PET is the main imaging modality allowing definition of the target tumor based on its metabolic properties [5] . For such oncological applications of FDG-PET, accurate tumor segmentation and estimation of the functional tumor volume are essential.
There are several problems for tumor segmentation on FDG-PET images. Although manual segmentation was often used because of its simplicity, it was time-consuming and susceptible to window level settings, and intra- and inter-observer variability [1] [6] . To overcome these problems, automatic and semi-automatic tumor segmentations have been studied, and several methods have been proposed. Zaidi et al. divided these techniques into four categories: 1) image thresholding methods, 2) variational approaches, 3) learning methods, and 4) stochastic modeling-based techniques [6] . Although using these techniques allowed the tumor to be segmented more accurately than with manual segmentation, they suffered from some problems specific to PET. For example, the lower resolution of PET relative to CT renders segmentation challenging, reducing the contrast between objects, with boundaries between the objects in PET images often being unclear. Another problem relates to image processing and smoothing filters. Noise is inherently high in PET images, and thus, image processing and smoothing filters are commonly utilized to reduce it. However, these can obscure tumor boundaries and make it difficult to segment the tumor accurately.
To overcome these PET issues, this study proposes a new method of image segmentation based on graph cut [7] . The proposed method efficiently utilizes local spatial information to construct the segmentation energy of graph cut, enabling accurate tumor segmentation. For comparison, three other techniques were also evaluated: region- growing based on 35%, 40%, and 45% of the tumor SUVmax, a region-based active contour model [8] , and simple linear iterative clustering (SLIC) superpixels [9] . The present paper was based on our previous study presented at SPIE medical imaging 2015 [10] .
2. Material and Methods
This retrospective study was approved by the institutional review board of our institution. The acquisition of informed consent was waived by the review board.
2.1. Patients and FDG-PET Images
Sixty-four bone tumors from 17 patients (11 men and 6 women; mean age, 61 years; range, 21 - 88 years) were included in this study. These bone tumors were clinically suspected to be metastases, and could be detected on FDG-PET images.
For each patient, whole body FDG-PET/CT images from the ear to the midthigh were acquired with a combined PET/CT scanner (Discovery 690; GE Healthcare, Waukesha, WI, USA). The PET component of the scanner can acquire 47 transaxial PET images in one bed position, with the following parameters: matrix size, 192 × 192 pixels; interslice spacing, 3.27 mm. The technical parameters for the CT component were: 16-detector row helical CT scanner; CT image matrix size, 512 × 512 pixels; interslice spacing, 3.27 mm. Patients received an intravenous injection of 160 - 320 MBq of FDG after at least 4 hours of fasting. About 60 minutes after the injection, low-dose CT was performed at a normal expiration position for attenuation correction of PET images. Then, an emission PET scan was performed at 2-min acquisition per bed position using a 3-D acquisition mode and time of flight method. Attenuation-corrected PET images were reconstructed with a 3-D ordered-subset expectation maximization reconstruction algorithm. In this study, only the PET images were analyzed.
2.2. Ground Truth for the Tumor Segmentation
To obtain the ground truth for the tumor segmentation, each bone tumor was manually delineated by a consensus between two board-certified radiologists (MN and AKK), who were also certified in diagnostic nuclear medicine. To reduce the computational cost, a 3-D bounding box surrounding the entire tumor was manually set by the radiologists. The radiologists evaluated the SUVmax of the tumor and the coordinates of the voxel where SUVmax was obtained (“the SUVmax voxel”). The FDG-PET images inside the bounding box, the SUVmax of the tumor, and the SUVmax voxel were used in the tumor segmentation. Pre- and post-processing were not utilized.
2.3. Graph Cut and Locally Connected Conditional Random Fields
In graph cut, image segmentation is performed via energy minimization [7] . One advantage of graph cut is that the characteristics of images and the target tumor can easily be incorporated into the segmentation energy. Another advantage is that graph cut can be applied to N-dimensional images. In our study, 3-D FDG-PET images were analyzed using graph cut. The segmentation results from graph cut are robust to noise; hence, in the medical field, this technique has been applied to the segmentation of anatomical structures [11] - [13] and to tumor segmentation with FDG-PET [14] [15] .
Generally, the segmentation energy of graph cut is divided into two terms, the unary term and the pairwise term. The energy E of the graph cut is represented as follows:
 (1)
(1)
where k is a constant that controls the balance between the unary and pairwise terms; V is the set of all voxels within the PET images; l is a label (0 represents non-tumor and 1 represents tumor); U(v, l) is the unary energy term when assigning a label l to voxel v; N(v) is the set of neighborhood voxels around voxel v; and P(v, w) is the pairwise energy term, which is small when the similarity between voxel v and w is high. Using graph cut, the segmentation energy E can be minimized and labels can be assigned to the set of voxels V.
In grid conditional random fields (CRF), a 6- or 26-connected neighborhood is commonly used as N(v), and the segmentation energy represented by Equation (1) can be minimized by using graph cut. In this study, locally connected CRF (LCRF) was proposed as the pairwise term. Compared with the 6- or 26-connected neighborhood, LCRF incorporates local spatial information into the segmentation energy more widely, allowing more reliable tumor segmentation. In LCRF, a 3-D cubic window with length L was set for each voxel, and voxels within the window were considered for the pairwise term. The N(v) of LCRF was represented as the following equation:
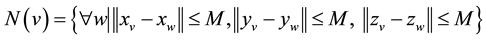
where 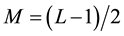 because L is assumed to be odd, and xv, yv, zv and xw, yw, zw are the x, y, and z-axis coordinates of voxels v and w, respectively. Figure 1 shows a schematic illustration of the set of neighborhood voxels, N(v), used in the pairwise term. As P(v, w), the commonly used energy function was used as follows:
because L is assumed to be odd, and xv, yv, zv and xw, yw, zw are the x, y, and z-axis coordinates of voxels v and w, respectively. Figure 1 shows a schematic illustration of the set of neighborhood voxels, N(v), used in the pairwise term. As P(v, w), the commonly used energy function was used as follows:
 ,
,
where Iv and Iw are the SUV of voxel v and w, and dist(v, w) is the Euclidean distance between voxel v and w.
For the unary term, an adaptive threshold was used. First, the Otsu threshold [16] was applied to the PET images, and each voxel was classified as either a foreground voxel or background voxel on the basis of the Otsu threshold results. Next, an adaptive threshold for the unary term was calculated according to the following equation:
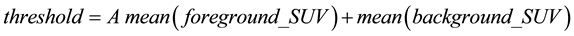 ,
,
where A is a parameter; and the mean(foreground_SUV) and mean(background_SUV) are the SUV means for the foreground and background voxels. The unary term U(v, l) is then defined as follows:

![]()
![]() (a) (b)
(a) (b)![]()
![]() (c) (d)
(c) (d)
Figure 1. Schematic illustration of the set of neighborhood voxels for the pairwise term of graph cut. When using graph cut, the neighborhood voxels, N(v), are determined in each voxel v. A 6- or 26-connected neighborhood is commonly used as N(v) in graph cut: (a) shows the 6-connected neighborhood; (b)-(d) show the n(v) of locally connected conditional random fields (LCRF) at L = 3, 5, and 7, respectively. The 26-connected neighborhood is identical to the LCRF when L = 3. In (a)-(d), 6, 26, 124, and 342 pairs are considered for the pairwise term, respectively.
2.4. Other Segmentation Methods
To compare the LCRF, three other types of segmentation methods were also applied: region-growing based on 35%, 40%, and 45% of the tumor’s SUVmax (RG35, RG40, and RG45, respectively), a region-based active contour model [8] , and simple linear iterative clustering (SLIC) superpixels [9] .
2.5. Region Growing
Region growing was performed for tumor segmentation using the SUVmax of the tumor and the SUVmax voxel as inputs. The seed point for region growing was the SUVmax voxel. The voxels were considered as candidates to be segmented if the SUV value of the voxel was greater than a lower threshold defined as a given percentage of SUVmax (in this study 35%, 40%, or 45%).
2.6. Region-Based Active Contour Model
Several methods have been suggested as a region-based active contour (AC) model. In our study, the Selective Binary and Gaussian Filtering Regularized Level Set method was used, which can efficiently determine the contours even if the boundary of the tumor is weak or blurred [8] . In addition, when using this method, the boundaries of a tumor can be detected automatically wherever the initial contour is located in the image. In our study, the initial contour was set at the SUVmax voxel.
2.7. SLIC Superpixels
Using SLIC superpixels (SS), sets of pixels can be divided into superpixels, which represent meaningful local structures [9] . Originally, SS used the 5-D space defined by the values L, A, and B of the CIELAB color space and the x and y pixel coordinates. In our study, 4-D space defined by the value of SUV and the x, y, and z voxel coordinates was used, and 3-D FDG-PET images were divided into several superpixels (supervoxels). After performing SS, the supervoxel that included the SUVmax voxel was regarded as the tumor segmentation. Merging of supervoxels was not performed in our study.
2.8. Parameters for the Segmentation Methods
In the current study, the following parameters were used for the segmentation methods:
1) RG35, RG40, and RG45 have no parameters.
2) SS has two parameters: superpixel compactness and the desired number of superpixels. Superpixel compactness ranged from 1 to 10, and the desired number of superpixels ranged from 1 to 20.
3) AC has two parameters: standard deviation of the Gaussian filter and the increase ratio of the level set function when solving the differential equation numerically. The standard deviation of the Gaussian filter ranged from 0.01 to 0.5. The increase ratio of the level set function ranged from 5 to 35.
4) In LCRF, there are three parameters: A of the unary term, L of the pairwise term, and k for controlling the unary term and pairwise term. The range of A was 0.3 - 0.7; that of L was 3 - 9; and that of k was 1 - 50.
2.9. Statistical Analysis
To validate the accuracy of tumor segmentation, a Dice similarity coefficient (DSC) was calculated between the manual segmentation and the result of each technique. DSC is one of the most widelyused quantitative metrics to evaluate segmentation accuracy [17] . DSC is defined as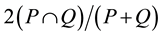 , where P is the result of manual segmentation and Q is that of the segmentation technique. The DSC ranges in value from 0 to 1, with 0 indicating no spatial overlap and 1 indicating a complete overlap between the two sets. Differences in DSC were tested using the Wilcoxon signed-rank test. R-3.0.3, a language and environment for statistical computing (available at http://www.R-project.org), was used to perform all statistical analyses. The significance level was set at p ≤ 0.05.
, where P is the result of manual segmentation and Q is that of the segmentation technique. The DSC ranges in value from 0 to 1, with 0 indicating no spatial overlap and 1 indicating a complete overlap between the two sets. Differences in DSC were tested using the Wilcoxon signed-rank test. R-3.0.3, a language and environment for statistical computing (available at http://www.R-project.org), was used to perform all statistical analyses. The significance level was set at p ≤ 0.05.
3. Results
The DSCs of the segmentation techniques are shown in Table 1. In each method, the optimal DSC value was selected from the combinations of the parameters. Table 1 also presents the results of the Wilcoxon signed-rank test comparing the LCRF at L = 9 with each other method. As shown, the best mean DSC (DSC = 0.812) was obtained with LCRF at L = 9. The DSC difference between LCRF at L = 9 and LCRF at L = 3 was statistically significant (p = 0.0151). Because LCRF at L = 3 is identical to the grid CRF with a 26-connected neighborhood, this result demonstrates that the DSC of the LCRF was better than that of the conventional grid CRF. Table 1 also shows that LCRF at L = 9 was significantly superior to the other methods: RG35, RG40, RG45, AC, and SS. Representative FDG-PET image and tumor segmentation results are shown in Figure 2, which demonstrates that the bone tumor was reliably segmented using LCRF at L = 9.
![]()
Table 1. Dice similarity coefficients between each type of image segmentation method and the manual segmentation, given as a mean and standard deviation across the 64 tumors.
Notes: p-values were results obtained from the Wilcoxon signed-rank test between LCRF at L = 9 and each of the other methods.
![]()
Figure 2. FDG-PET image and tumor segmentation results for an 84-year-old man with a metastatic bone tumor of the right ischium. (a) FDG-PET image, (b) manual segmentation, (c) LCRF at L = 9, (d) AC, (e) SS, (f) RG40. Abbreviations: LCRF: locally connected conditional random field; AC: region- based active contour model; SS: SLIC superpixels; RG40: region-growing based on 40% of the tumor maximum standard uptake value.
4. Discussion
In this study, we proposed a new segmentation method for FDG-PET images based on graph cut. This method was applied to the clinical FDG-PET images of patients with metastatic bone tumors. The results of our study show that tumor segmentation was reliably performed using LCRF, and that the performance of LCRF was better than those of the other segmentation methods. In LCRF, the local spatial information was efficiently incorporated into the pairwise term of the segmentation energy, which led to accurate tumor segmentation.
Tumor segmentation and the determination of the tumor boundary on FDG-PET images are needed for quantitative evaluation of FDG-PET and guidance for localized cancer therapies (such as radiation therapy and interventional radiology procedures). Apart from manual segmentation, thresholding methods were the most widely used approach in clinical practice for delineating tumors on FDG-PET images. However, sensitivity to noise and inability to handle intensity variations made it difficult to perform tumor segmentation robustly in clinical cases using thresholding methods [16] . A number of alternative methods have been proposed. Among them, Ballangan et al. used graph cut to segment lung cancer on FDG-PET images [14] . Compared with the previous method, the novel advantage of our method was that LCRF can, at least potentially, be combined with any form of U(v, l) and P(v, w), such as the downhill SUV feature proposed by Ballangan et al.
Table 1 shows that LCRF at L = 9 was superior to that of the grid CRF with 26-connected neighborhood for tumor segmentation (which is identical to LCRF at L = 3). Because U(v, l) and P(v, w) in Equation (1) are identical in both LCRF at L = 9 and the grid CRF with 26-connected neighborhood, the difference in the performance was due to the number of neighborhood voxels incorporated into the pairwise term of the equation. This result suggests that local spatial relationship is useful for accurate tumor segmentation. In contrast to LCRF, the use of local spatial information was limited in the segmentation processes RG35, RG40, RG45, and AC. In RG35, RG40, and RG45, tumor segmentation was affected by the connectivity to the seed point. In AC, the output of the level set function is affected by the image gradient and the means of all tumor and non-tumor voxels. We speculate that the segmentation result of LCRF is better than those of RG35, RG40, RG45, and AC because the local spatial information could be utilized efficiently in LCRF.
SS generates superpixels based on clustering of the local neighborhood [9] . Thus, both SS and LCRF utilize local spatial information for image segmentation. However, the distance measure for clustering in SS does not reflect the image feature specific to FDG-PET images because it has been designed for general purpose. Because the unary and pairwise terms of LCRF can be specialized in the segmentation on FDG-PET images, LCRF surpassed SS for tumor segmentation on FDG-PET images.
The transition from the tumor to background can be gradual on FDG-PET images, which makes it difficult to segment a tumor accurately. AC, SS, and LCRF can handle this problem by using local or global information from PET images. However, when the SUV distribution of tumor was heterogeneous, the segmentation results using LCRF were more accurate than those of AC and SS, as shown in Figure 2. This demonstrates that tumor segmentation with LCRF was more robust than with AC and SS.
The use of automatic or semi-automatic tumor segmentation has rarely been studied in bone tumor. To our knowledge, only Torigian et al. have evaluated the thresholding methods to segment spinal bone metastases on FDG-PET images [18] . In the current study, the segmentation targets were not limited to spinal bone, and the advanced segmentation technique was applied to clinical FDG-PET images. In metastatic bone tumor, heterogeneity in tumor SUV can frequently be observed because osteolytic and osteoplastic lesions can be combined in one tumor. In addition, the shape of a metastatic bone tumor is often not spherical as metastatic bone tumor invades surrounding tissue along the shape of the bone. Because of these two points, it is difficult to segment a metastatic bone tumor accurately, and the number of studies using automatic or semi-automatic segmentation on FDG-PET images is limited. Because metastatic bone tumors are often treated with radiation therapy, the segmentation of bone tumors will be important for PET-guided radiation therapy.
There are several limitations to the current study. First, the numbers of patients and tumors included in this study were small or moderate. For further study, it is necessary to confirm the robustness and applicability of our segmentation method in a clinical setting by using a larger cohort of patients. Second, a widespread major problem for validating an image segmentation method is construction of the ground truth, and this problem was also found in the current study. Although it is highly desirable to validate the segmentation methods with pathology-based ground truth [19] [20] , errors are often introduced due to changes in tissue morphology during excision, fixation, and slicing [19] . In addition, metastatic bone tumor is often diagnosed clinically without pathological evaluation. Therefore, at present it is difficult to use pathology-based ground truth for validation of bone tumor segmentation. If precisely co-registered PET images and pathological images become available, we will validate our segmentation method using the pathology-validated PET images. Third, we did not apply our methodology to co-seg- mentation in PET/CT images. Han et al. [15] used graph cut in order to segment head and neck cancer on FDG- PET/CT images, and segmentation accuracy was improved compared to the graph cuts method solely based on PET images by using both the PET and the CT information. We expect that LCRF can be applied to co-seg- mentation in PET/CT images and improve accuracy in the co-segmentation method.
5. Conclusion
A new segmentation method based on graph cut was proposed for FDG-PET images. By using LCRF, local spatial information was efficiently incorporated into the segmentation energy of graph cut. Our results demonstrated that the results of LCRF surpassed those of conventional grid CRF, and that a tumor was more reliably segmented with LCRF than with other techniques.
NOTES
*Corresponding author.