1. Introduction
Ganoderma lucidum (G. lucidum), a fungus famous as traditional Chinese herbal medicine, has been widely used for preventing and treating a series of diseases. G. lucidum spores are the fungus’s reproductive cells ejected from the cap of G. lucidum after its fruiting body becomes mature. Though the fruiting body of G. lucidum has been widely utilized as a Chinese medicine for several thousand years, the spores of G. lucidum have been realized and utilized only since the 20th century. Recent studies demonstrated that the spores of G. lucidum not only inherit all active ingredients of G. lucidum, but also have stronger bioactivities, about 75 times more than G. lucidum’s fruiting body regarding its effect, such as enhancing immunity, antitumor, preventing diabetes, protecting liver and so on [1] [2] . As G. lucidum spores are valuable Chinese herbal medicine, its potent bioactivity and wide acceptability make it an important marketable product. However, the market product from fruiting body of G. lucidum sometimes pretends to be product from G. lucidum spores for economic benefit. Therefore, it is of great importance to develop an accurate, efficient and simple method to identify G. lucidum spores from its fruiting body since it is very difficult to discriminate them from their market products.
G. lucidum spores have a complicated system of compounds. The commonly investigated methods for the analysis of herbal medicines, like high performance liquid chromatography (HPLC), thin layer chromatography (TLC) and colorimetry, are found to be expensive, time-consuming, labour-intensive, and requiring large quantity of organic solvents. Also, the results are inadequate for classification purpose because of the limited amount of active chemical components that can be detected in what is a very complex system in G. lucidum spores [3] [4] .
Attenuated total reflection Fourier transform infrared (ATR-FTIR) spectroscopy is very efficient for revealing chemical compositions and structures of herbal medicine in terms of easy and direct usage of technique, non- destructiveness, small quantity of sample needed and short data acquisition time. Since chemical processing of the herbal material is not needed at all, the chemical composition of the material remains in its original form [5] . However, studies on herbal medicines using the ATR-FTIR technique are still in its early stages [6] and there were few studies on ATR-FTIR spectroscopy of G. lucidum spores.
ATR-FTIR spectra of herbal medicines consist of many overlapping absorption bands representing the different modes of vibration of a large number of molecular constituents in the compounds. These vibrational bands are sensitive to the physical and chemical states of the compounds, and they can be detected at low levels [4] . However, the differences in the ATR-FTIR spectra within the same herbal species may be subtle and it is difficult to distinguish between spores and fruiting body of G. lucidum through simple visual inspection. Thus, approriate multivariate chemometric methods have been applied in this study to analyze the ATR-FTIR spectra for feature selection and classification. Although the spectroscopic data are highly correlated, often, only a small subset of spectral features is found essential. Due to the complex nature of the combined spectroscopic data, it is of great interest to determine if a small subset of spectral reflectance measurements contains as much informative feature for classification purpose as the whole spectrum does. There is little scientific research paper available regarding the spectral feature and chemical composition of G. lucidum spores.
In this paper a penalized discriminant analysis (PDA) model was developed to identify informative spectral features for distinguishing between spores and fruiting body of G. lucidum. The multivariate methods using principal component discriminant analysis (PCDA) and partial least squares discriminant analysis (PLSDA) were also explored based on the whole spectrum. The model performances based on the selected wavelength bands and the whole spectrum were compared in terms of classification accuracy and interpretation of spectral features. The established discrimination models explored in this paper would be an accurate, simple and robust tool for the quality control of G. lucidum spores. In particular, the discriminant vectors can be helpful for the interpretation of spectral features needed for discrimination and for providing a quantitative explanation of the major chemical constituents of G. lucidum spores regarding its anti-cancer effects.
2. Materials and Methods
2.1. Sample Preparation
Ten samples of G. lucidum fruiting body and ten G. lucidum spores samples were originated from Taishan, China. The fruiting body of G. lucidum was cross-sectioned into thin slices. From each sample, multiple spectra were taken from five different positions: top surface, middle area, bottom surface, outer stipe and inner stipe. In total, 80 spectra from G. lucidum fruiting body samples and 30 spectra from G. lucidum spores samples were collected.
2.2. Spectral Acquisition
A Fourier transform infrared (FTIR) spectrometer (Perkin-Elmer Spectrum 100 model) with an attenuated total reflectance (ATR) accessory was used to record the absorbance spectra of the G. lucidum fruiting body and spores directly without any processing. The ATR-FTIR spectra of all the G. lucidum samples were recorded in the mid-IR region of 4000 - 400 cm−1 at resolution of 4 cm−1 with 20 scans for each spectrum. Each spectrum with high signal-to-noise signal of about 50 was obtained by an average of these 20 scans. Background spectra were always recorded before running the sample spectra in order to obtain absorbance spectra with smooth baseline and with minimum detection of absorption bands of water vapor and carbon dioxide present in the optical path of the infrared beam in the spectrometer. Strong absorption bands in the absorbance spectra of the G. lucidum samples were accurately obtained by applying sufficient pressure on the sample onto the ZnSe crystal using a diamond tip in the ATR set-up [5] .
2.3. Spectral Pre-Treatment
ATR-FTIR spectra are affected by both the concentration of the chemical constituents and the physical properties of the analyzed product. The physical effects, such as baseline variation, light scattering, path length differences, etc., account for the majority of the variance among spectra while the variance due to chemical composition is considered to be small. Therefore mathematical pretreatments are essential to reduce the variation due to physical effects so as to enhance the contribution of the chemical composition [7] [8] .
The spectra were first smoothed using the Savitzky-Golay algorithm [9] by spanning a 10-point window, and then were reduced by taking every sixth point to speed up subsequent manipulation. To remove the regions of the spectra with low signal-to-noise ratios arising from the lower system response, only the wavenumbers ranging from 4000 to 450 cm−1 with 593 spectra points were used in the analysis. To remove slope variation and to correct light scatter due to different particle sizes, the standard normal variate (SNV) method [10] was used to standardize each spectrum by setting its mean intensity to zero and the variance to one. After pretreatment, the mean spectra from fruiting body and spores of G. lucidum have similar patterns as shown in Figure 1, which indicates similar chemical composition. Some small between-class differences were observed in the region of 1800 - 1050 cm−1, 3000 - 2700 cm−1 and 3700 - 3500 cm−1, which may indicate different levels of the chemical components contained in fruiting body and spores of G. lucidum.
![]()
Figure 1. Mean spectra of fruiting body (solid line) and spores (dotted line) of G. lucidum after spectral pre-treatment.
2.4. Statistical Analysis
1) Principal component discriminant analysis (PCDA) and partial least squares discriminant analysis (PLSDA)
Multivariate statistical methods including principal component analysis (PCA), partial least squares (PLS) and linear discriminant analysis (LDA) [11] - [13] were employed on whole spectral region to investigate the differences of spectra from spores and fruiting body of G. lucidum.
Simple and direct implementation of LDA in high-dimensional spectroscopic data setting provides poor classification results and the interpretation of the results is challenging due to singularity problem and highly-correlated spectral features. To solve this problem, many traditional approaches to this problem involve performing feature selection to reduce the variable dimension before classification. PCA and PLS were used to reduce the dimension of the original spectral data matrix with little loss of information. Then LDA focuses on finding a linear combination of the new variables, provided either by PCA or PLS, to construct canonical variate which best separates the two groups. Using pretreated spectral data described in Section 2.3, classification rules were derived using principal component discriminant analysis (PCDA) [14] - [16] and partial least squares discriminant analysis (PLSDA) [15] - [17] . The PCDA involves an initial PCA on the pre-treated spectra followed by a LDA performed on the first k PCs’ scores. The PLSDA involves a PLS regression on the pre-treated spectra followed by a LDA on the first k PLS components’ scores. Both PCDA and PLSDA were carried out with k ranging from 2 to 20.
2) Penalized discriminant analysis (PDA)
Though the spectroscopic data are highly correlated due to the presence of a large number of overlapping broad peaks, in many applications, only a small subset of variables (wavelengths/wavenumbers) contain sufficient information for discrimination. Hence there is interest in determining if a small subset of spectral wavenumbers containsas much information as the full spectrum does.
Penalized discriminant analysis (PDA) [18] [19] involves a sparse linear combination of all spectral features by imposing an 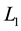 norm penalty on the discriminant vector. The PDA extends LDA to the high-dimensional setting to deal with ill-posed problem without filtering out any spectral feature. Using pretreated spectral data with number of spectra points p = 593, classification rules were derived by a PDA using Fisher’s criteria [20] . It generates discriminant vector v in Fisher’s discriminant problem which involves a sparse linear combination of p number of spectral features by shrinking some coefficients towards zero and setting others equal to zero. The PDA investigated here can be expressed in the following optimization problem:
norm penalty on the discriminant vector. The PDA extends LDA to the high-dimensional setting to deal with ill-posed problem without filtering out any spectral feature. Using pretreated spectral data with number of spectra points p = 593, classification rules were derived by a PDA using Fisher’s criteria [20] . It generates discriminant vector v in Fisher’s discriminant problem which involves a sparse linear combination of p number of spectral features by shrinking some coefficients towards zero and setting others equal to zero. The PDA investigated here can be expressed in the following optimization problem:
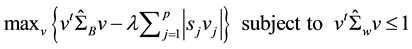
where 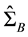 is the sample estimate of between-class covariance matrix and
is the sample estimate of between-class covariance matrix and 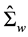 is the sample estimate of within-
is the sample estimate of within-
class covariance matrix, λ is non-negative tuning parameter. A diagonal estimate for the within-class covariance matrix is used here because it has been shown to give good results in the high dimensional setting [21] . Each feature of the discriminant vector v is additionally penalized by its sample within-group standard deviation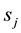 . The larger the value of λ, the larger the penalty on v and therefore the smaller the number of non-zero components, that is, the resulting discriminant vector v will be sparse in the spectral features. Classification accuracy was used to guide the choice of optimal parameter.
. The larger the value of λ, the larger the penalty on v and therefore the smaller the number of non-zero components, that is, the resulting discriminant vector v will be sparse in the spectral features. Classification accuracy was used to guide the choice of optimal parameter.
Leave-one-out cross-validation was used to train the algorithm by carrying out the PCDA, PLSDA and PDA classification rules on all the data except one sample which was then tested. This was repeated until all samples have been tested and an overall model accuracy was determined. To ensure that the wavelengths selected by PDA model are not training set specific, the PDA model was also validated with 70% of the data being treated as training data and 30% as test data. To ensure the statistical robustness, this process was repeated 50 times with different random splits of training and test sets, and the average misclassification rates were presented to assess the classification performance.
All the algorithms for computations and analyses were implemented in R statistical programming language [22] .
3. Results and Discussion
3.1. Absorption Band Assignments of ATR-FTIR Spectra of G. lucidum
Figure 1 shows the typical ATR-FTIR spectrum of G. lucidum after pretreatment in the region of 4000 - 450 cm−1 with major peaks of the absorption bands labeled on the mean spectrum. The spectral wavenumbers and their corresponding assignments of the absorption bands in the ATR-FTIR spectrum of G. lucidum were given in Table 1 based on literature [7] [23] - [26] .
Polysaccharide, triterpene, sterols, amino acids, proteins, fatty acids have been known as the most biologically active substances in G. lucidum spores [27] [28] . The bioactive polysaccharides in the forms of glucomannan and arabinan isolated from G. lucidum spores, identified by the absorption band at 1064 cm−1, 1035 cm−1 respectively (listed in Table 1), have been demonstrated to exhibit strong immunomodulation and anti-tumor activities including preventing oncogenesis and tumor metastasis [23] [24] . Furthermore, synergistic effect of polysaccharides and other bioactive components such as triterpene and sterols compounds isolated from spores G. lucidum, identified in the absorption band at 1415 cm−1, 1377 cm−1 and 1145 cm−1 (given in Table 1), have shown to possess high bioactivities and proved effective as cytotoxic, antiviral and antiinflamatory agents [25] [27] . In the regions like ~1630 cm−1, 1733 - 1710 cm−1, 2957 - 2852 cm−1, spectra receive contributions from the compounds, such as protein, fatty acids and lipids of G. lucidum spores [25] [27] .
3.2. Discrimination by PCDA and PLSDA
For PCDA and PLSDA model using the full spectrum region, the number of PCs or PLS components chosen is crucial to the discrimination performance. The discrimination results of cross-validation were used to optimize the number of PCs or PLS components. For PCDA model, the first seven PCs were used to construct the discrimination model and the leave-one-out cross-validation analysis gave a discrimination accuracy of 97%. With the relationship between the spectra variables and the responses taken into account for latent variable design, the PLSDA model used a fewer optimal number of latent variables (only three PLS components) when constructing the canonical variate and the leave-one-out cross-validation achieved a discrimination accuracy of 99%. In Figure 2 the 3D scatter plot of the first three PLS components illustrated a very clear separation between spores and fruiting body of G. lucidum in a 3D space.
![]()
Table 1. Absorption band assignments [7] [23] - [26] of the ATR-FTIR spectrum of G. lucidum.
![]()
Figure 2. The 3D scatter plot of the first three PLS components’ scores of the spectra from G. lucidum spores (○) and its fruiting body (▲).
3.3. Discrimination by PDA
PDA model seeks to find an optimal parameter λ with lowest error rate. Figure 3 shows the relationship among the tuning parameter λ, the number of the selected non-zero features (representing the sparsity level of the discriminant vector) and the corresponding error rate of PDA model. It illustrates that the sparsity level of the solution varies smoothly with λ, with larger values of λ resulting in very sparse solutions. When is less than 0.11, larger values of λ result in sparser solutions and lower error rate. The drop is observed at around 50 non-zero features. When λ is more than 0.11, larger λ gives higher error rate. The penalized LDA model with optimal tuning parameters of λ = 0.11 and 53 (out of 593) selected wavelengths points gives classification accuracy of 99% for discrimination between spectra of spores and fruiting body of G. lucidum. The contributions of this wavelength selection method are essential. It leads to a model having a small number of selected wavelength regions, yet comparable or better classification ability than full-spectrum model. Out of 50 random splits of data, the number of times the wavelength was selected shows how important is the wavelength to the discrimination. The selected wavelengths recorded in Figure 4 are concentrated in the regions of 3700 - 3500 cm−1, 3000 - 2800 cm−1, ~1700 cm−1, ~1400 cm−1, ~1000 cm−1, which are consistent with the spectral variation in Figure 1. The selected wavelengths show us which parts of the spectrum are important to the discrimination.
It is possible that the superior performance of PDA model relative to the models using full wavelength is due to the fact that ATR-FTIR spectroscopic data consist of many overlapping absorption bands of which only a small proportion may be informative for explaining the response. Including those uninformative wavelength points in a model may introduce a great deal of noise and thus reduce the performance of the model.
The good discrimination results from all these models suggested that there may exist some inherent compositional differences between spores and fruiting body of G. lucidum.
3.4. Correlation between Spectral Absorption Bands and Chemical Components of G. lucidum and Its Medicine Effect
Discrimination performance of the models may be explained by the correlation between spectral features and chemical constituents of G. lucidum spores. With non-zero features selected for discrimination, the discriminant vector of PDA model makes direct and valuable contribution to interpreting spectral features related to the medical
![]()
Figure 3. Left panel: Number of non-zero features versus tuning parameter; Right panel: Misclassification rate versus selected non-zero features.
![]()
Figure 4. Wavenumbers selected for discrimination between spores and fruiting body of G. lucidum. The height of the bars shows the number of times the wavenumber was selected in 50 random splits of the data.
effect of G. lucidum spores. In Figure 5, the informative features selected are in the regions of 1150 - 980 cm−1, 1700 cm−1, 3000 - 2700 cm−1 and 3700 - 3500 cm−1 corresponding to some differences between the mean spectra of spores and fruiting body of G. lucidum. The non-zero regions with prominent absorption features around 1150 - 1050 cm−1 are typical features of triterpenoids and polysaccharides due to the C-O vibrations, which are the major chemical constituents of G. lucidum. The other prominent absorption peak selected at around 1700 cm−1 is consistent with a C=O stretching vibration in carbonyl compounds which may be characterized by the presence of high content of terpenoids and protein in G. lucidum. It is reported that G. lucidum spores possess a much higher content of triterpenoids on a weight basis when compared to G. lucidum fruiting body [29] . The selected features with sharp peak at around 3000 - 2700 cm−1 are due to C-H stretching vibration. The non-zero regions of 3700 - 3500 cm−1 are characteristic of carbohydrates proteins due to the O-H stretching vibration as shown in Table 1. The carbohydrate content in fruiting body of G. lucidum is much higher than that in spores [27] - [29] , which also explains the mean spectra differences between fruiting body and spores of G. lucidum in this region.
For the PCDA and PLSDA model, the PCDA or PLSDA loading of the original variables combines the loading from PCA or PLS and the loading from LDA when constructing a canonical variate. Therefore the PCDA loading and the PLSDA loading show the contribution at each wavelength to the linear diagnostic rule and thus can be related easily to the spectral features, which permits interpretation of its spectral basis. A comparison between PCDA loading and the PLSDA loading for discrimination between spores and fruiting body of G. lucidum can be seen in Figure 6. The loading features emphasized by the PLSDA model are very similar to those
![]()
Figure 5. Mean spectra from G. lucidum spores (red line) and its fruiting body (blue line) with PDA coefficients in green for optimal tuning parameter.
![]()
Figure 6. Comparison of PCDA loadings (blue line) and PLSDA loadings (green line) for discrimination between the spectra from fruiting body and spores of G. lucidum.
emphasized by PCDA model, which are consistent with group differences between spores and fruiting body of G. lucidum as shown in Figure 7. When comparing the PDA coefficient in Figure 5 with PCDA and PLSDA loadings in Figure 6, we found that the non-zero regions with prominent absorption features from PDA model were also observed as the most prominent features by PCDA loadings and the PLSDA loadings. However, for the PLSDA and PCDA model, when the number of components used for the discrimination is getting bigger, the loadings from these two models may become more complex and thus the contribution of each wavelength to the classification becomes less interpretable when it is related to the spectral features. The major features of these loadings can also be explained by the assignments of the corresponding absorption bands in the ATR-FTIR spectrum listed in Table 1. The high consistency between the selected features by the discrimination models and chemical features of the ATR-FTIR spectrum may provide a quantitative explanation of the major chemical constituents of spores G. lucidum with respect to chemometrics.
G. lucidum contains approximately 400 different bioactive compounds [22] . Among these ingredients, triterpenoids, polysaccharides, sterols, proteins, nucleo-sides and fatty acid are found the major chemical constituents of G. lucidum [28] . These biologically active compounds have been demonstrated to play a significant role in prevention of oncogenesis and tumor metastasis [6] . The spores of G. lucidum possess a much higher amount of bioactive substances than the fruiting body of G. lucidum [28] . In particular, polysaccharides from G. lucidum
![]()
Figure 7. PLSDA loadings for discrimination between the spectra from spores and fruiting body of G. lucidum. The PLSDA loading is shown in green, with the mean spectra for the two types superimposed (fruiting body in blue line, spores in red line).
spores have immunomodulating properties. G. lucidum spores have been found to be effective in modulating the immune responses, and thus show efficacy of immunostimulatory and antitumor activities. Some comparative studies also reported that spores and fruiting body of G. lucidum showed different efficacy with regard to their antitumor effects and immunomodulatory activities [1] [29] .
4. Conclusions
The spores of G. lucidum have higher economic value compared with its fruiting body due to the fact that numbers of bioactive substances of the spores are much higher than those of the fruiting body of G. lucidum. Since a variety of commercial G. lucidum products are available in various forms, it is of essential importance to distinguish spores and fruiting body of G. lucidum for the purpose of quality assurance.
In this study, the combination of ATR-FTIR spectroscopy and chemometrics method has been proved to be a very powerful tool to distinguish G. lucidum spores from its fruiting body efficiently. An excellent classification performance of up to 99% accuracy can be achieved by the discrimination models using the spectral features either selected or emphasized by the proposed models. By imposing penalties on the discriminant vectors, the PDA model presented in this paper enables an automatic selection of a small number of informative wavelength points to construct an efficacious discrimination model, which gives comparable or even higher accuracy than the PCDA and the PLSDA models based on the full wavelength.
Most essential contribution of the model is that the selected spectral regions for discriminant analysis show a good link between spectral features and chemical components of G. lucidum spores, which provided some evidence for its anticancer effect. This is a novel and important finding, as it provides quantitative interpretation and scientific support to the claims on the health benefits and antitumor properties of G. lucidum spores. It is also a potentially useful tool for quality control and fast discrimination of raw materials of traditional herbal medicine. Identified spectral regions may be targeted for further analysis linked with its active biochemical components of herbal medicine.
Acknowledgements
This research is supported by Academic Research Funds (AcRF: RI 12/10 TTL and AcRF: RI 6/14 ZY) of National Institute of Education, Nanyang Technological University, Singapore.
NOTES
*Corresponding author.