Automatic and Manual Proliferation Rate Estimation from Digital Pathology Images ()
1. Introduction
The development and continued growth of cancerous cells involve various changes at both macro and micro levels of the body. Cell proliferation is usually among the major indicators for proliferation of cancerous cells. Specifically, breast cancer proliferation rate estimation (PRE) is a crucial step for determining the cancer level and is used as a prognostic indicator [1] . In conjunction with tumor size and grade, lympth node status and histological grade, PRE is an indicator for the aggressiveness of individual cancers and helps setting the treatment plan [2] .
Traditionally, pathologists perform proliferation rate estimation for breast cancer by examining the whole slides via a microscope. Over the past two decades, digital pathology enabled the usage of high resolution digitizers to provide high resolution images that replace the microscope as shown in our previous work [3] .
There are many clinically approved techniques to estimate the PRE including: mitotic index, S-phase fraction, nuclear antigen ImmunoHistoChemistry (IHC) including KI-67 and PCNA-staining Cyclins and PET [4] [5] . Each one of these methods has its advantages or disadvantages based on the clinical settings.
In our work, we use Ki-67-stained biopsy images for PRE. In this technique, PRE is estimated by counting the number of brown nuclei and the number of blue nuclei as shown in Figure 1. Stromal areas are clinically excluded from counting because stromal area does not become cancerous. In our previous work [6] , we performed digital stromal area removal to eliminate this ambiguous area for both junior pathologists and automated PRE systems.
Manual PRE is time-consuming and laborious for pathologists. An average of six minutes per image is required for PRE by an expert pathologist. Our expert pathologist requires over 10 hours estimating the proliferation rate for our dataset containing 100 images. Many authors target automation of PRE including our recent work [7] . However, one major concern was not investigated in all these efforts which was the inter-variability between the expert pathologists [8] .
In this paper, we study the statistical inter-pathologist variability for the various manual PRE we have between four expert pathologists. Moreover, we investigate the reliability of our proposed automated PRE compared to the four pathologist opinions for the 100 images in our dataset.
2. Materials and Methods
Manual ground truth estimation is a major area of interest due to the various human factors that influence the experts. Specifically for breast cancer PRE [9] [10] , we find that pathologists provide variable ground truth estimations which make it hard to evaluate any automated PRE estimated technique. Many automated PRE techniques have been proposed in the literature and we recently proposed our technique in [7] , an exhaustive review of the techniques as well as a detailed description of our techniques are presented in [7] . In this paper, we provide the necessary statistical study for the inter pathologists variability. Furthermore, we study the statistical variability between the four manual ground truth and our automated technique. In [7] , we compared the automated results with one expert pathologist and a student trained by a pathologist. In this paper, we run our statistical study to include for expert pathologists and one automated technique.
We study three statistical significance tests to show the inter-observer variability. Moreover, we study the manual vs automated [7] PRE variability. We study three statistical significance measures: correlation coefficient, T-Test, and Ch-Square test. We briefly describe them due to space limitations.
2.1. Correlation Coefficient
The value of the correlation coefficient 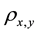 gives an indication about the strength and the nature of the relationship between two random variables x and y. It ranges between −1 and +1. A value of +1 means perfect correlation between these two random variables while a −1 values indicates maximal uncorrelated variables [11] the equation for the correlation is shown in Equation (1):
gives an indication about the strength and the nature of the relationship between two random variables x and y. It ranges between −1 and +1. A value of +1 means perfect correlation between these two random variables while a −1 values indicates maximal uncorrelated variables [11] the equation for the correlation is shown in Equation (1):
![]()
Figure 1. The sample images of Ki-67 stained pathology images showing sample blue nuclei, and stromal areas.
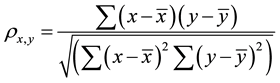 (1)
(1)
where x and y are two random variables, 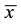 and
and  are the corresponding mean values for each sample.
are the corresponding mean values for each sample.
2.2. Student T-Test
Student T-Test (or t test for short) is one of a number of hypothesis tests. The t-test looks at the t statistic, t distribution and degrees of freedom to determine a t value (probability) that can be used to determine whether the two underlying distributions of the two random variables are different as shown in Equation (2):
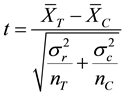 (2)
(2)
where 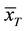 and
and 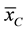 are the two mean values for the corresponding two data samples r and c,
are the two mean values for the corresponding two data samples r and c, 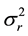 and
and  are the corresponding variances for the two data samples r and c, nT and nC are the number of the corresponding samples. Moreover, the degrees of freedom (df) for the test should be determined. In the t-test, the degree of freedom is the sum of the persons in both groups minus 2. Given the alpha level, the df, and the t value, you can look the t value up in a standard table of significance Typically, when t > 0.05, the two random variables (two underlying data samples) are said to be statistically insignificance, i.e., highly correlated [12] .
are the corresponding variances for the two data samples r and c, nT and nC are the number of the corresponding samples. Moreover, the degrees of freedom (df) for the test should be determined. In the t-test, the degree of freedom is the sum of the persons in both groups minus 2. Given the alpha level, the df, and the t value, you can look the t value up in a standard table of significance Typically, when t > 0.05, the two random variables (two underlying data samples) are said to be statistically insignificance, i.e., highly correlated [12] .
2.3. Chi-Square
Chi square X2 is a statistical test commonly used to compare observed with unobserved data upon a specific hypothesis as in Equation (3):
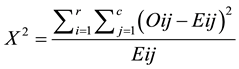 (3)
(3)
where Oij is the observed frequency in the ith row and jth column, Eij is the expected frequency in the ith row and jth column, r is the number of rows and c is the number of columns. The appropriate number of degrees of freedom (df) is calculated as the number of rows-1 multiplied by the number of columns. If X2 is greater than what is known as the critical value, then the two samples are dependent.
3. Experimental Results and Analysis
Our data set contain 100 Ki-67-stained histopathology digital images for breast cancer. The blue nuclei are negative positive cells while the brown nuclei are the positive ones. Our collaborating pathologists provided us with the ground truth from four different pathologists including herself as the most senior pathologist. We provided each pathologist with anonymized images labeled in sequence along with an sheet to score for the blue and brown nuclei. None of the four pathologists knew about the other and they were scoring independently. Our most senior pathologist (coauthor) spent over 10 hours for scoring the 100 cases which means an average of 6 minutes per case. Moreover, we run our proposed automated PRE system proposed in [7] over the same 100 images and recorded the automated scoring for both the blue and the brown nuclei.
3.1. Correlation Coefficient
The inter-observer reproducibility is first measured by using the correlation coefficient [13] [14] . Overall, there is a higher correlation between pathologists in brown nuclei estimation than blue nuclei estimation. Moreover, our automated CAD system has also a higher correlation coefficient for brown nuclei compared to blue ones. Table 1 summarizes the inter pathologists correlation coefficient values and manual vs automated correlation coefficient values.
From Table 1, we note that the correlation coefficient indicates a very high correlation between the four observers on the brown nuclei counting. However, the correlation is highly variable for blue nuclei counting from an upper value of 0.73 down to 0.768. Figure 2 and Figure 3 show the relationship for the manual PRE for
![]()
Table 1. Significance values of correlation coefficients.
![]()
Figure 2. Relationship between first and second observers’ nuclei count estimates.
![]()
Figure 3. Relationship between third and fourth observers’ nuclei count estimates.
observer 1 vs observer 2 and observer 3 vs observer 4, respectively.
On the other hand, we study the correlation coefficients between the manual of each of the four experts and our proposed automated system as shown in Table 2. As we examine this table, the brown nuclei counting is highly correlated to the various observers which indicates an almost perfect reliability of our proposed automated system for brown nuclei estimation. Furthermore, the blue nuclei counting are comparable to the correlation between the manual observers. In other words, our automated blue nuclei estimation is as good as the manual estimation which proves its clinical reliability.
3.2. T-Test
We performed a two-tailed paired T-Test on all the pairs between the four observers and the automated system. Our Null Hypothesis is that there is a difference between the observers in one hand and the automated system on the other hand. All of the reported significance probability values in Table 3 shows insignificant statistical difference between the manual expert estimations themselves on one hand and between both the 3rd and 4th observers with the automated system on the other hand. In other words according to Table 4 which shows the interpretation for the p-value. As you see the p value is less than 0.01 which means that we have a strong evidence to reject the hypothesis that says that there is no relationship (there is a difference) between observers on one hand and the automated system in the other hand in both Brown and Blue nuclei counts estimation.
3.3. Chi Square
We computed Chi-square test all pairs, and it compared with the critical chi square value with df = 1, confidence level 99% (probability = 1 − 0.99 = 0.01). In all pairs (including inter-observer and our automated method), the calculated chi square value is greater than the critical value, which means that each pair of samples are dependent. In other words, it is statistically reliable to consider any of the expert scoring or the automated scoring values. Figure 4 and Figure 5 show two samples images where we high agreement between observes, and a low agreement between observers, respectively.
![]()
Table 2. Manual vs automated significance values of correlation coefficients.
![]()
Table 3. Significance values resulting from paired T-Test.
![]()
Table 4. Interpreting significance probability (p-value).
![]()
Figure 4. Example of an image has the same value for the brown nuclei in all observers.
![]()
Figure 5. Example of an image where the observers results are completely different.
4. Conclusion
We proposed a detailed statistical study for breast cancer proliferation rate estimation. We studied the inter- observer variability between four expert pathologists on a set of 100 cases. We also studied the reliability of our recently proposed automated PRE system. On the 100 cases, we found that the variability of brown nuclei estimation was statistically insignificant between various pathologists. We also found that our proposed system brown nuclei estimation was statistically reliable. On the other hand, our three statistical significance tests showed fairly high reliability between pathologists for blue nuclei estimation. The same conclusion applies for our proposed automated blue nuclei system.