1. Introduction
Being constituents of important primary metabolites, nitrogen and sulphur represent for all plant cells essential macronutrients, hence their deficiency triggers a wide range of metabolic responses in higher plants and microalgae. To investigate about the effects of mineral nutrients shortage in plant cell, microalgae represent a valuable support for their rapidity of growth, simply culture system and high reproducibility of experiments. Therefore, it is necessary to distinguish the effects due to a long-time starvation from those deriving from a short-time starvation. At this regard it should be pointed out that what for an higher plant is a starvation of short duration in the context of hours, for algae it should be considered a long-time starvation. Metabolic changes associated with nutrient deprivation in the green microalgae Chlorella sorokiniana occurred in a time-dependent manner, generally reaching a maximum in the first day (24 h) of starvation, but depending on the specific growth rate in any organism. In this context, a Chlorella sorokiniana suspension that contained all nutrients in sufficient concentration showed a growth constant of 3 d−1.
In microalgae, long-term nutrient deprivation can lead to the cell death preceded by autophagy, a self-de- grading process to recycle part of the cytoplasm including organelles [1] .
Although nutritional deficiencies determine in algal cells common adaptation strategies, we have previously demonstrated that N- or S-starved cells of Chlorella sorokiniana display different metabolic trends, suggesting that different response mechanisms exist to compensate for the absence of these two nutrients. As a macroelement, N has a profound importance for microalgae metabolism and its limitation is compensated by radical changes in metabolic pathways. N-deprivation stimulates de-repression of enzymes involved in N metabolism such as nitrate reductase [2] , and 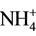 or
or  transport systems [3] [4] . In addition, in the process of acclimation to long term N-deficiency, microalgae have been reported to degrade ribosomes and decrease enzyme activities involved in gluconeogenesis and photosynthetic carbon fixation cycle [1] . These effects of N-starvation recall in many aspects those observed following S-deprivation. Indeed, S-starvation causes de-repression of enzyme systems involved in sulphate transport, metabolism and cysteine synthesis [5] - [7] . In addition, microalgae have been reported to synthesize arylsulphatase to recover
transport systems [3] [4] . In addition, in the process of acclimation to long term N-deficiency, microalgae have been reported to degrade ribosomes and decrease enzyme activities involved in gluconeogenesis and photosynthetic carbon fixation cycle [1] . These effects of N-starvation recall in many aspects those observed following S-deprivation. Indeed, S-starvation causes de-repression of enzyme systems involved in sulphate transport, metabolism and cysteine synthesis [5] - [7] . In addition, microalgae have been reported to synthesize arylsulphatase to recover  [8] . Such changes represent attempts to obtain N or S from environment by highly specific uptake systems and to assimilate them even if their external concentrations are very low.
[8] . Such changes represent attempts to obtain N or S from environment by highly specific uptake systems and to assimilate them even if their external concentrations are very low.
In this study, the authors determine and compare, in C. sorokiniana, the effects of N- or S-starvation on C metabolism and evaluate the importance of N in the sulphur assimilation and of S in the nitrogen assimilation. In particular, GS and OASTL, key enzymes in N and S assimilation pathways respectively, were taken into consideration. The enzyme GS consents the insertion of inorganic nitrogen into glutamate to form the amide glutamine, while the enzyme OASTL inserts inorganic sulphide into OAS to synthesize cysteine. Many papers indicate these two enzymes are finely regulated in plant cell [7] [9] .
The aim was also to clarify aspects of the interaction among N, S and C metabolisms in a photosynthetically active plant cell. Besides, these findings may contribute to the knowledge of the metabolic consequences to mineral deprivation in microalgae.
2. Materials and Methods
2.1. Algal Growth Condition
All experiments were performed by using Chlorella sorokiniana Shihira & Krauss, strain 211/8K (CCAP of Cambridge University). The algae were grown in batches placed in a thermostatic chamber at 35˚C, continuously stirred and illuminated by fluorescent lamps (150 µmol photons m−2∙s−1). The cultures were insufflated with air containing 5% CO2 at a flow rate of 80 - 100 l∙h−1. Three different types of medium were used to grow cells: basal, nitrate-free and sulphate-free medium. The composition of the basal medium was previously reported [10] . S- and N-starved cells were obtained collecting batch grown cells by a low speed centrifugation (4000 r for 5 min), washed two times and re-suspended in a N-free or S-free medium as previously described [3] [11] . Algal growth was evaluated by absorbance (OD550). Culture purity was monitored by optical microscopy.
2.2. PCV Determination
The packed cell volume (PCV) was estimated by centrifuging 10 ml of cell suspension in a haematocrit tube at 4000 r for 5 min. At the end of centrifugation the PCV value can be directly read from the calibrated tube.
2.3. Glutamine and Cysteine Determination
Intracellular glutamine concentration was tested in sufficient and in S- or N-starved cells. Cell suspensions (10 ml) were collected by centrifugation (4000 r for 5 min), the packed cells mixed with a solution containing 1 ml cold 80% ethanol and 1 mM γ-aminobutyric acid, left for 15 min and then centrifuged (4000 r for 5 min). The supernatant was filtered through Waters Sep-Pak® Cartridges to remove chlorophyll. The glutamine was determined by HPLC as previously described [12] . For cysteine content determination the packed cells were treated with 2 ml of extraction buffer as previously described [12] . Cysteine content was quantified by reversed-phase HPLC after derivatization with monobromobimane according to Carfagna et al. [12] .
2.4. Glutamine Synthetase (GS) and O-Acetylserine(thiol)lyase (OASTL) Assays and Cysteine Determination
To prepare crude extract for GS activity determination, aliquots of 100 ml of algal culture were harvested by centrifugation at 4000 r for 5 min; the pellets were re-suspended in 5 ml of extraction buffer ( 10 mM Tris-HCl pH 7.2, 2.5 mM MgCl2, 1 mM dithiothreitol, 5 mM ethylenediaminetetraacetic acid, 15% glycerol) and the cells were lysed by a passage at 11,000 psi through French pressure cell (Aminco). To 1 ml of mix reaction ( 40 mM imidazole-HCl pH 7.0, 30 mM glutamine, 20 mM K-arsenate pH 7.0, 120 mM NH2OH pH 7.0, 0.4 mM adenosine diphosphate, 3 mM MnCl2) 1 ml of crude extract was added. The whole mixture was incubated at 30˚C and after 10 min, 2 ml of stop mixture (4% FeCl3, 2.4% trichloroacetic acid, 0.6 M HCl) was included. Subsequent to a low speed centrifugation the absorbance of the mix was measured at 540 nm. One enzyme U is defined as 1 µmol of γ-glutamylhydroxamate min−1∙mg−1 protein. To determine OASTL activity, aliquots of 100 ml of cultures were harvested by centrifugation (4000 r for 10 min) and the pellets were re-suspended in the extraction buffer ( 50 mM phosphate-buffer pH 7.5, 10 μM pyridoxal phosphate and 1 mM dithiothreitol). The cells were broken by passing twice through a French pressure cell (11,000 psi). The lysates were clarified by centrifugation at 15,000 rpm for 15 min at 4˚C. The supernatant represented the crude extract. The enzymatic activity of OASTL was measured according to Gaitonde method [13] modified as described by Carfagna et al. [14] . One enzyme U is defined as 1 µmol cysteine min−1∙mg−1 protein. Protein amounts were determined using the Bio- Rad protein assay based on the Bradford method [15] with bovine serum albumin as the standard.
2.5. Starch Content Determination
Cells were collected by a low speed centrifugation (4000 r for 10 min). The packed cells were extracted twice with 80% ethanol at 80˚C for 15 min. After cooling, the pellets were re-suspended twice in distilled water and then centrifuged. The pellets were washed twice in 50 mM acetic acid-NaOH buffer (pH 4.8) by centrifugation and then autoclaved for 30 min at 120˚C. The extracts were cooled and total starch was determined as glucose derived from its hydrolysis as previously described [12] .
2.6. Total C, N and S Content Determination
2.7. Statistical Analysis
Experimental data analyses were made using Sigmaplot 12 software. All data are expressed as the means ± SE for 5 to 9 determinations. The statistical analysis was performed by one-way ANOVA with a Tukey post-hoc test to determine differences between sufficient and S or N starved algae, P < 0.05 and P < 0.001 as significant. If necessary, the data were log + 1 (x) transformed before the analysis. Data expressed in percentages were trans-
formed by arcsin transformation (y′ = arcsin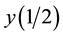 ,
,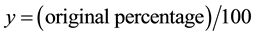 ) and then analyzed.
) and then analyzed.
3. Results and Discussion
3.1. Effects of Nutrient Deprivation on Algae Growth
In order to define and compare the effects of N- or S-starvation on algae growth, we performed OD550 determination. When cells were in the exponential phase of growth (OD550 about 0.8), they were starved of N- or S-nu- trient and the growth monitored for the following 48 h of nutrient deprivation (Figure 1 and Figure 2). Cells were incubated in culture media with a starting cell density of approximately 414.062 cells/ml.
Some previous papers reported reduction in microalgal growth rate and increase of cell volume as a consequence of long-term N- and S-starvation [1] [16] .
We observed that cellular growth is more affected in N-starved cells if compared to S-starved ones (Figure 2(b)).
S-deprivation implies a reduction of cell growth that is less evident than in N-starvation: the fact that in S- starved cells the rate of growth has a slowdown delayed probably corresponds to a period of cellular sulphur recycling. As previously reported in Chlorella sorokiniana cells [14] , upon S-deprivation, the degradation of intracellular glutathione has the purpose to maintain Cys homeostasis. Besides, this growth discrepancy between N- and S-starved cells is likely caused by the relative importance and abundance of N compared with S: while S can be obtained from intracellular stores as glutathione [14] , N must be supplied constantly for adequate growth because it cannot be sufficiently compensated by recycling pathways. In fact, nitrogen occurs in every protein and most metabolites and total N content of dry C. sorokiniana biomass is almost 10-fold greater than the total S content. As such, a much greater mass of N ( 10 mM KNO3) is necessary to facilitate growth of C. sorokiniana, while a comparatively lesser amount of S ( 1.2 mM MgSO4) is sufficient. In 24 h of nutrient shortage, significant changes in the average cell volume did not occur (Figure 1). The slight cell enlargement caused by N- depriva- tion is likely due to the starch store occurring in N-starved cells (Figure 1 and Table 1). S-starved cells appear small and grouped suggesting a likely presence of mucilage material (Figure 1).
3.2. Effects of Nutrient Deprivation on Metabolites Content and Enzyme Activities
Our studies showed that N-starvation generally yielded similar effects as S-starvation, but the impacts on cell growth and total protein were much more severe.
![]() (a)
(a)![]() (b)
(b)
Figure 1. Chlorella sorokiniana cultures growth under different condition. C: Algal cells growth in a basal complete medium; -S: Sulphur starved culture; -N: Nitrogen starved culture (Panel (a)). Optical Microscope images (100×) of sufficient (C) and S- or N-starved cells (Panel (b)).
![]()
Table 1. Starch content in Chlorella sorokiniana cells growth in sufficient, N- or S-starved medium.
![]()
![]() (a) (b)
(a) (b)
Figure 2. Growth of Chlorella sorokiniana 211/8K in basal complete medium (Panel (a)). When cells were in the exponential growth phase (OD550 about 0.8), they were starved of N or S and the growth monitored for the following 48 h of nutrient deprivation (Panel (b)).
Total soluble protein concentration in algal samples decreased drastically in response to nutrient starvation. A 65% decrease in protein content was observed after 24 h of S-deprivation and a decrease of over 80% was observed after 24 h of N-starvation (Figure 3). Typically, in microalgae, the decrease in protein content was simultaneous to the increase of starch (Table 1) and lipid upon nutrient starvation [16] [17] .
Strikingly, S-deficiency also affects free amino acids pool, which appeared greatly increased compared to sufficient algae [12] , while S-containing amino acids like Cys appeared at low level [14] . Among amino acids, glutamine, the first organic compound deriving from N-assimilation, increased following S-starvation and obviously resulted reduced in N-deprived cells (Figure 4). Noticeably, GS activity, which resulted enhanced in N- deprived cells, was not affected by S-deprivation (Table 2). The glutamine increase in S-starved cells could indicate an imbalance of N-assimilation and protein synthesis caused by the inadequacy of Cys or methionine. Furthermore, N-starvation affect intracellular Cys content. A reduction of 50% in cysteine intracellular level was observed after 24 h of N-starvation (Figure 4). In fact, the backbone for cysteine synthesis is O-acetylserine (OAS) which in turn derives from serine, a product of N-assimilation. The enzyme OASTL catalyses the synthesis of cysteine from OAS and sulphide and represents an important point of connection between the nitrogen and sulphur assimilation [7] [18] . Indeed, our results show that OASTL activities were strongly decreased in N- starved cells (Table 2). The enzyme OASTL exists in several isoforms that are localised into different compartments of the plant cell [19] [20] . S-deprivation induced OASTL activities increase in C. sorokiniana cell [7] [11] , similar to those observed in a number of higher plants [21] . The increase in OASTL activity during S-depriva- tion could be ascribed to an increase in the cytosolic OASTL isoform [7] [11] .
3.3. Changes in Total C, N and S under Nutrient Deprivation
N- or S-deprivation causes a loss of photosynthetic capacity and a decrease in the cells chlorophyll content. As previously reported, chlorophyll content strongly decreased upon both S- and N-starvation [3] [22] . Several studies indicate the degradation of PSII as responsible for this decrease [23] . Chlamydomonas reinhardtii is known to restructure its photosynthetic machinery upon S-deprivation to minimize oxidative stress that may occur in conditions of reduced photosynthesis [1] . As such, the decrease in chlorophyll content could be ascribed to the deactivation and disassembly of photosynthetic complexes as a response to oxidative stress resulting from S-starvation. As well, protein degradation that occurs in N-starved cells does not save the enzyme ribulose-1,5-
![]()
Figure 3. Contents of total soluble proteins in sufficient, and in S- or N-deprived cells for 24 h. The protein contents (expressed as μg∙μl−1 PCV) were determined by Bradford method. Data are presented as means ± SE (n = 5). Columns with the same letter were not significantly different (P ≤ 0.05, ANOVA, Tukey multiple comparison).
![]()
Figure 4. Contents of glutamine and cysteine in sufficient, and in S- or N-deprived cells for 24 h. The amino acids contents (expressed as μmol∙L−1 PCV) were determined by HPLC. Data are presented as means ± SE (n = 5). Columns with the same letter were not significantly different (P ≤ 0.05, ANOVA, Tukey multiple comparison).
bisphosphate carboxylase oxygenase [1] [24] and the depletion of this protein may compromise the mechanism of photosynthesis, leading to the decrease in chlorophyll content observed in N-deficient Chlorella cells.
Our previous results [3] [22] suggest that a global shutdown in energetic functions may occur upon N- or S- deprivation: photosynthesis and chlorophyll levels drop considerably, but also respiration resulted decreased.
Our recent investigation showed that activation of antioxidant enzymes in S-deprived C. sorokiniana occurred in a period of 24 h of S-starvation and that these increases correspond to the rapid raise in Reactive Oxygen Species (ROS) occurring in the first hour of starvation [25] .
In this paper we show that total C is reduced by 10% in N-deprived and 16% in S-deprived cells. Another interesting result is that in cells S-deprived the total N is reduced by only 23%, whereas in cells N-deprived the total S is reduced by 47%. These data seem to confirm that N-deprivation strongly affects S-assimilation (Figure 5).
![]()
Table 2. Glutamine synthetase and O-acetylserine(thiol)lyase activities in sufficient and N- or S-starved cells for 24 h.
![]()
Figure 5. Contents of total carbon (C), nitrogen (N) and sulphur (S), expressed as % dry weight in sufficient, and in N- or S-deprived cells for 24 h. Data are presented as means ± SE (n = 9). Columns with different letter were significantly different (P ≤ 0.05, ANOVA, Tukey multiple comparison).
It has also been shown that N-deprivation increases the C/N ratio (6.4), probably due to reduced synthesis of amino acids and proteins and to the storage of starch. In S-starved cells the C/N ratio slightly increased (4.7) respect to that of sufficient cells (4.3).
4. Conclusion
N- and S-deprivation influence C metabolism as photosynthetic activity and starch storing in a similar mode. Anyway, cellular growth is more affected in N-starved cells if compared to S-starved ones. S-deprivation implies a reduction of cell growth but with delay with respect to the N-starvation. Probably, the cellular recycling in response to nutrient deprivation is more effective under S-respect to N-shortage. In addition, the relative importance and abundance of N compared with S could explain this growth discrepancy between N- and S-starved cells. The total N content in C. sorokiniana cells is estimated to be almost 10-fold greater than the total S content. Our data show that N-deprivation powerfully affects S-assimilation. In N-deprived cells the total S is reduced by 47%, whereas in S-deprived cells the total N is reduced by 23%. The cysteine intracellular content decreased in N-starved cells by around 50% and OASTL activity resulted strongly reduced. Our data demonstrate that in Chlorella sorokiniana a mutual influence of N, S and C assimilation occurs.
NOTES
*Corresponding author.