Reference Genes for RT-qPCR Analysis of Environmentally and Developmentally Regulated Gene Expression in Alfalfa ()
1. Introduction
Cultivated alfalfa (Medicago sativa L.) is a major forage legume grown extensively worldwide [1] . This perennial species has a tetraploid (2n = 4x = 32) genome which displays tetrasomic inheritance and its populations are extremely polymorphic due to a high degree of outcrossing [2] . Superior tolerance to environmental stress and decline in stem digestibility during plant development are major issues for alfalfa breeding programs [3] . Insufficient tolerance to subfreezing temperatures reduces long term persistence of alfalfa in northern countries and is a source of major costs associated with stands reestablishment and the need to buy forage outside the farm [4] . Lignin deposition in maturing stems significantly interferes with the release of simple sugars from the cellulose and hemi-cellulose fiber fraction and causes a significant decline in feed value [5] . The development of new breeding approaches allowing significant gains will require a deeper knowledge of these complex traits at the genetic and molecular levels. Analysis of candidate gene expression within heterogeneous populations will help identify genes and gene networks that affect these traits. It will also facilitate the search for trait-allele associations in projects pursuing high throughput assessment of DNA polymorphisms.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR) is a highly sensitive technique which can detect small differences in transcript abundance between samples even for weakly expressed target genes. Normalization of cDNA concentrations with stably expressed genes is needed to minimize the effects of uncontrolled factors such as variation in RNA concentrations or the efficiency of reverse transcription that can affect the accurate determination of transcript variations across biological samples [6] . The so-called “reference genes” are often selected for their constitutive expression or their involvement in maintenance of cell structure or in primary metabolism. Genes encoding 18S ribosomal RNA (18S rRNA), actin (Act), tubulin (Tub), glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and eukaryotic elongation factor 1-alpha (eEF1-α) are often used as putative “housekeeping genes” even though they may not always meet the requirements for internal controls [7] [8] . It has been well documented that the selection of reference genes can markedly affect the accuracy of RT-qPCR analysis and that no single gene or genes combination should be arbitrarily used without validation under specific experimental conditions and varied sources of genetic material [9] . For that reason, reference gene candidates have been recently tested in a wide array of plant species and experimental conditions [10] - [21] . These studies clearly demonstrate the need to validate reference gene candidates under the experimental conditions specific to each study in order to avoid erroneous conclusions. Several statistical approaches have been developed to select reference genes [7] . The geNorm M value based on the analysis of pairwise variations between reference genes is a widely used indicator of the expression stability of candidates [22] .
In this study we evaluated the stability of the expression of several candidate reference genes in different genetic backgrounds of cultivated alfalfa exposed to variable environmental conditions or sampled at different developmental stages. Our objective was to identify genes that are stable under a wide range of conditions and would constitute good candidates for the selection of reference genes in RT-qPCR analysis of the expression of genes potentially associated to key agronomic traits in alfalfa. We also assessed the impact of unstable reference genes for normalization of gene expression in the assessment of their relationship with physiological traits.
2. Material and Methods
2.1. Plant Materials
Plants of alfalfa (Medicago sativa spp. sativa) were sampled during their acclimation to various environmental conditions (low temperature, water deficit, photoperiod) and at different stages of their development. These plants were obtained from a series of independent experiments described below:
2.1.1. Cold Acclimation Indoors
Plants of the cultivars Apica (ATF0) and Evolution (ETF0) and populations (ATF5 and ETF5) obtained after five cycles of recurrent selection for superior tolerance to freezing (TF) within these two cultivars [23] were grown and cold-acclimated under environmentally-controlled conditions. Plants were grown from seeds in 15-cm pots (10 plants per pot) filled with a mixture (10:3; v:v) of top soil/peat moss (Pro-mix BX, Premier Peat Moss, Rivière-du-Loup, QC, Canada) supplemented with a controlled release fertilizer [N: 17% (w:w); P: 7.31% (w:w); K: 14.1% (w:w); 250 g/35 L; Muticote 4, Haifa Chemicals Ltd, Haifa Bay, Israel]. Plants were grown six weeks in an environmentally-controlled chamber set to: photoperiod, 16 h; day-time temperature, 22˚C; night- time temperature 17˚C. Artificial lighting was provided by a mixture of high pressure sodium and metal halide 400 W lamps (PL light systems, Beamsville, ON, Canada) with a photosynthetic photon flux density (PPFD) of 600 to 800 µmol photons m−2∙s−1. Plants were kept well watered and fertilized twice a week with a 1 g∙L−1 of a commercial fertilizer (20-20-20 plus micronutrients, Plant-Prod, Brampton, ON, Canada). Plants were subsequently cold acclimated at constant 2˚C, 8-h photoperiod with 150 µmol photons m−2∙s−1 PPFD. After two weeks at 2˚C, plants were transferred to a freezer set to −2˚C in the dark for a second stage of hardening to promote superior freezing tolerance.
Two separate cold acclimation assays were performed: “cold acclimation 2011” and “cold acclimation 2013”. In 2011, non-acclimated (NA) plants of populations ATF0 and ATF5 were sampled immediately before their transfer to low temperature and at the end of the cold acclimation period at −2˚C (A). In 2013, the populations ETF0 and ETF5 were added and a time course analysis of gene expression was performed by taking samples 0 h, 8 h, 1 d, 3 d, 7 d and 15 d after transfer to 2˚C and after two additional weeks of hardening below freezing at −2˚C (HF). In both experiments, crown tissues of four randomly selected pots (10 plants per pot) of each population were harvested at each sampling point.
2.1.2. Cold Acclimation Outdoor
Plants of the initial cultivars ATF0 and ETF0 and populations ATF5 and ETF4 obtained after respectively five and four cycles of recurrent selection were grown under environmentally-controlled conditions and acclimated to natural hardening conditions in an unheated greenhouse as described in Castonguay et al. [24] . Five pots (15 plants per pot) of each population were sampled before transfer the unheated greenhouse on October 15, 2003 and during the subsequent winter on January 21, 2004 after fall hardening.
2.1.3. Water Stress and Photoperiod
Two additional treatments were included in the “cold acclimation 2013” experiment to test candidate reference genes under water deficit and reduced photoperiod. For that purpose, unstressed plants of ATF0 and ATF5 grown under the conditions described previously (22˚C/17˚C, 16 h photoperiod) were compared to plants 1― Exposed to a progressive water deficit by withholding water during a 10 d period until the appearance of wilting symptoms; 2―Maintained under short (8 h) photoperiod at warm temperature (22˚C/17˚C) for three days. Five replicates (10 plants∙pot−1) were available for each treatment.
2.1.4. Plant Development
Plants of the biomass-type cultivar Orca were grown in a greenhouse under the conditions described in Duceppe et al. [25] . Stems with leaves were harvested from 20 genotypes with high and 20 genotypes with low cell wall degradability at three developmental stages (vegetative, full bloom and mature pods). At each harvest, a single stem was collected from each genotype, chopped into 3 cm segments and flash frozen in liquid N2. Stems of each group of genotypes were pooled into bulk samples for RNA extraction.
2.2. Primer Design
Reference gene candidates tested in this study are listed in Table 1. Except for Actin primers that were developed from a partial Medicago sativa coding sequence available in GenBank (EU664318), other target sequences for candidate genes were developed from an alfalfa EST library (Serge Laberge, unpublished) prepared from cDNAs from cold-acclimated crowns (pooled RNA extracts from 50 genotypes) of the population ATF2 obtained after two cycles of recurrent selection for superior tolerance to freezing within the cultivar Apica [23] . EST sequences with functional homologies with genes that show expression stability in other plants [12] [14] or that maintained a stable expression in a macro array hybridization with cDNA from non-acclimated and cold- acclimated plants of the alfalfa population ATF2 (Serge Laberge, unpublished) were used to design primers with the Oligo Explorer software, version 1.1.0 (T. Kuulasma, University of Kuopio, Kuopio, Finland; Table 1). Selected EST sequences have been deposited in the GenBank data library under the names and associated accession numbers listed in Table 1.
2.3. RNA Extraction and cDNA Synthesis
RNA samples were extracted from crowns with buds (5-cm transition zone between shoots and roots) or stems
![]()
Table 1. Candidates as reference gene (Ref) and genes of interest (GOI) with primer sequences, PCR fragments characteri- stics and amplification efficiency. Selection criteria (literature vs macro array hybridization) and GenBank accession number of Medicago sativa EST used to design primers are also provided.
†Setection based on literature; ‡Setection based on differential hybridization of alfalfa cDNA library.
with leaves and ground to a fine powder in an analytical mill (IKA A11, Wilmington, NC) equipped with a cutting blade and a reinforced chamber for embrittlement of tissues in liquid N2. Total RNA was extracted using a CTAB procedure as described in Dubé et al. (2013). Total RNA was quantified using the ExperionTM RNA StdSens microcapillary chip (Bio-Rad, Mississauga, ON, Canada) and its integrity based on the RNA quality indicator (RQI) calculated by the ExperionTM software was always greater than 8. First-strand cDNA was synthesized from 1 µg of total RNA and oligo(dT)18 primers using the Transcriptor First Strand cDNA synthesis Kit (Roche Applied Science, Laval, QC, Canada) following the manufacturer instructions. Any residual genomic DNA was removed by a treatment with DNaseI (Invitrogen, Burlington, ON, Canada) prior to cDNA synthesis. Two independent cDNA synthesis reactions were performed for each sample and used as technical replicates in RT-qPCR analyses.
2.4. Real-Time PCR
RT-qPCR analysis of gene expression was performed according to the MIQE guidelines [6] . Assays were carried out as described in Dubé et al. [26] in a Mastercycler® ep realplex system (Eppendorf Canada, Mississauga, ON, Canada) using the QuantiTect® SYBR GreenPCR kit (QIAGEN, Toronto, ON, Canada). The 10-µl reaction mixture contained 3 µl of first-strand cDNA and 0.5 µM of each of the forward and reverse primers. The thermocycler program was set to: 15 min activation at 95˚C followed by 40 cycles of 15 s at 95˚C, 15 s at 60˚C; 1 min extension at 72˚C. Real-time PCR was carried out as duplicates and control samples without template or with RNA alone without reverse transcription were included as checks for potential contamination with genomic DNA. Twenty (20) µl of each reaction were run on 2% agarose gels stained with ethidium bromide to check the specificity of amplification. The DNA fragments were recovered using the QIAquick gel extraction kit (QIAGEN Inc., Mississauga, ON, Canada) and confirmed by sequencing. PCR efficiency (%) was calculated from the linear regression of a seven fold dilution of each PCR product using the following equation: Efficiency % = (10(−1/slope) − 1) × 100. The threshold cycle (Cq) values at which the PCR product fluorescence rises over the background fluorescence was determined by the instrument software which was set to default parameters. Data was analyzed with the qBasePLUS software version 3.0 (Biogazelle, Ghent, Belgium). Inter-run calibrators were included on each plate to account for plate-to-plate variation. Normalized relative expression levels was calculated using the 2−ΔΔCq or comparative Cq method based on the differences in Cq between the target and the reference genes and corrected for PCR efficiency [27] .
2.5. Statistical Analysis
Statistical analysis of gene expression data was performed with the two-way Anova procedure of SigmaPlot® Ver. 12.0 (Systat Software, San Jose, CA). Data normality was verified using the Shapiro-Wilk statistic. Pairwise comparison of means was performed with the Tukey test and statistical significance was postulated at P > 0.05.
3. Results and Discussion
3.1. Selection of Candidate Genes for Normalization of Gene Expression
Reverse transcription quantitative PCR (RT-qPCR) is a highly sensitive technique that has become the standard for gene expression analysis [28] .Various factors including variation in RNA integrity, reverse transcription efficiency or amount of cDNA templates between samples are significant sources of uncontrolled errors in RT- qPCR analysis of gene expression [9] . The normalization of samples against two or more reference genes is therefore required to account for technical variation and to achieve reliable estimates of true biological effects [9] [29] . Expression levels of reference genes in RT-qPCR analyses can be extremely variable under different experimental conditions. Consequently, a systematic validation of the stability of expression needs to be performed in order to identify the most stable genes in a given experiment and to determine the optimal combination required for normalization [9] . In that perspective a validated repertoire of genes stably expressed in distinct genetic backgrounds of cultivated alfalfa exposed to variable environmental conditions or sampled at different developmental stages would be a valuable resource for functional analyses of genes associated to key agronomic traits in that species.
We evaluated twenty one (21) candidates as potential reference genes for normalization of transcript levels in RT-qPCR analysis of gene expression in plants of alfalfa exposed to low temperature (Table 1). The selection of candidate genes was based on the following criteria:1―Previous reports of expression stability under various environmental conditions and different species [12] [14] or; 2―Lack of differential expression in a comparative macroarray hybridization of an EST collection with cDNA from non-acclimated and cold-acclimated crowns of alfalfa (Table 1). High amplification efficiencies that varied between 91 and 99% were observed for all selected candidates. This indicated an efficient annealing of the primers allowing an accurate detection of Cq and robust RT-qPCR assays [11] . Some sequences have previously been validated as reference genes in alfalfa [30] [31] and other plant systems [12] [14] . Other candidates including a vacuolar H+-ATPase A subunit (ATPase), a coatamer delta subunit (COP), cafeic acid 3-O-methyl transferase (COMT), an inositol transporter (ITR), profiling 1 (Pro1) and a retention in reticulum 1 protein (Rer1) homolog that we selected on the basis of the stability of their expression in a differential macroarray hybridization had not been previously tested as candidates.
3.2. Stability of the Expression of Candidate Reference Genes
The stability of the 21 candidate reference genes expressed as the geNorm mean pairwise variation (M) of a gene in comparison to the other genes was first tested with samples from plants acclimated to low temperature under controlled conditions in 2011 (Figure 1(a)). Using a M threshold value of ≤ 0.5 [27] , the most stably expressed genes were 18S rRNA, UBL-2a, ADF, CAC, ATPase, Rer1, eF-1α, COP, UBQ-5, eIF-2, UBQ-2 and GAPDH. Conversely Act, COMT and Tub were deemed unstable under these conditions. An M value just below the 0.5 geNorm threshold for GAPDH is also noteworthy. Act and Tub were shown to be unsuitable as references to normalize the expression of genes in sweet potato (Ipomoea batatas (L.) Lam) samples exposed to cold [16] . Graphical depiction of the number of cycles required for the fluorescence signal from amplified fragments to rise above the background level (Cq values) for two variable (Act and Tub) and two stable (UBL-2a and 18S rRNA) candidate genes in the alfalfa populations ATF0 and ATF5 illustrate the contrasted stability of these two set of genes (Figure 2). With their expression levels significantly repressed in plants exposed to low temperature (higher Cq values), Act and Tub clearly fall short of the reference genes requirement to maintain a stable expression regardless of the experimental conditions. This is in stark contrast with the highly stable expression of UBL-2a and 18S-rRNA under these experimental conditions. Act has been previously shown to be unstable under various experimental conditions and was deemed unsuitable to normalize stress-induced gene expression in several species [12] [21] [32] including alfalfa [30] . Rapacz et al. [18] concluded that the stability of Act in barley (Hordeum vulgare) exposed to abiotic stress could depend on the developmental stage. Tub is another gene commonly used for normalization of gene expression that has been frequently shown to be unstable across species and experimental treatments [9] [12] [16] [20] [30] .
The 12 candidate genes below the 0.5 M threshold in Figure 1(a) were further evaluated for their stability in response to environmental, developmental and genetic variations (Figure 1(b)). This extensive analysis tended to confirm the initial observations with regard to the stability of these candidates except for18S rRNA which was more unstable in the other data sets (Figure 1(b)). A much higher geNorm M value for 18s rRNA in the 2013 cold acclimation experiment indicates an instability as a reference gene even for replicated experiments conducted under identical conditions. Xia et al. [32] recently noted that 18S rRNA was unstable as a reference gene (geNorm M value ≥ 0.5) in coconut (Cocos nucifera L.) partly because of the abundance of its transcripts. It has been pointed out that large differences in the levels of target and reference gene transcripts may affect the accuracy of RT-qPCR analyses [11] . On the other hand, UBL-2a, ATPase and Rer1 were highly stable across a wide range of experimental conditions and are strong candidates as reference genes in RT-qPCR studies of gene expression in alfalfa. Ubiquitin protein ligase genes were selected as the most suitable reference genes across RNA samples from various tissues of Brazilian rubber trees (Hevea brasiliensis) treated with plant growth regulators [14] . It should be noted that Rer1 and ATPase genes that were included as candidates on the basis of their stability of expression in a differential macroarray hybridization of alfalfa ESTs had not been previously tested as putative reference genes in other species. New reference genes identified by microarray analysis in Arabidopsis thaliana [33] and soybean (Glycine max) [34] were shown to outperform commonly used housekeeping genes in these species and other plant systems as well [21] . It is also noteworthy that eEF-1α consistently maintained an M value well below the 0.5 geNorm threshold across all treatments and genetic sources and is another candidate to consider in the validation of reference genes in RT-qPCR analysis of gene expression in alfalfa. This is in agreement with previous reports of eEF-1α stable expression in various species tissues and treatments [10] [32] [35] including in low temperature-treated crowns of barley [36] .
![]() (a)
(a)![]() (b)
(b)
Figure 1. (a) Expression stability estimated with the geNormPlus M value for 21 candidate reference genes in the 2011 cold acclimation experiment. Candidates with lower M values have a more stable expression across all sampling points and replicates. (b) Twelve candidate genes with M values below the 0.5 cut-off threshold in panel (a) were subsequently tested for the stability of their expression under various experimental conditions including: 1―Repeat of the 2011 cold acclimation protocol in 2013; 2―A time course of cold acclimation with two cultivars (Apica and Evolution); 3―Exposure to a short photoperiod at high temperature; 4―Water stress and; 5―Sampling at different developmental stages or in plants of contrasting stem degradability. Bold numbers indicate reference gene with lowest values under each experimental condition.
Data in Figure 1(b) also reveal that even though GAPDH is frequently used as a housekeeping gene in RT-qPCR studies, it seldom met the requirements for robust analysis of gene expression in our study. The poor performance of GAPDH as a reference gene in cold acclimation experiments is corroborated by the significant induction of its expression in plants of four populations of alfalfa exposed to low temperature (Figure 3). This is
![]()
Figure 2. Quantification cycle (Cq) values of four candidate reference genes in a RT-qPCR assessment of expression in samples from the 2011 cold acclimation experiment. The Cq value is the number of cycles needed to rise above the threshold fluorescence signal and reflects the abundance of transcripts present in a sample. Each bar represents the mean of four replicates for non-acclimated (NA) and cold-accli- mated (A) plants of populations ATF0 and ATF5. For each gene, treatments with the same letter are not statistically different at P > 0.05.
![]()
Figure 3. Relative quantification of GAPDH expression during a time course of cold acclimation within cultivars Apica and Evolution and TF5 populations obtained after five cycles of recurrent selection for superior freezing tolerance within these two genetic backgrounds (TF0). Samples were taken 0 h, 8 h, 1 d, 3 d, 7 d and 15 d after transfer to 2˚C and after two additional weeks of hardening below freezing at −2˚C (HF). The validated reference genes used for normalization were Rer1 and ATPase. Each bar represents the mean of four replicates. Within each population, relative expression values with the same letters are not statistically different at P > 0.05.
in accordance with previous reports of its poor performance as a reference gene in other species [10] [19] including the normalization of gene expression in cold-treated plants of lentil (Lens culinaris) [19] and coconut [32] .(Zhu et al. 2013)(Saha and Vandemark (2013)) GAPDH and Tub were recently ranked by geNormPlus as among the most unstable candidate for the normalization of gene expression in alfalfa [30] . However, conflicting reports on the stability of the expression of GADPH and considerable regulation of eEF-1α in plants of the legume Cicer arietinum exposed to abiotic stresses illustrate that generalization should be avoided in the selection of reference genes and that the optimal combination must be validated under each specific experimental conditions. {Castro, 2012 #2710}{Castro, 2012 #2710}{Castro, 2012 #2710}{Castro, 2012 #2710;Castro, 2012 #2710;Castro, 2012 #2710@@author-year}{Abberton, 2008 #2154}{Abberton, 2008 #2154}{Garcia de Castro, 2000 #791}{Castro, 2012 #2710}{Castro, 2012 #2710}This task will be facilitated in alfalfa by the identification of strong candidates in the current study.
A combination of reference genes is required to ensure the accuracy of RT-qPCR analysis of gene expression [22] . However, increasing the number of internal controls is costly and time consuming and they should be kept to the minimum required for accurate normalization. The pairwise variation analysis of the optimal number of reference genes determined as the geNorm V value was used to evaluate the benefits of adding more reference genes on the accuracy of the normalization factor. The value is based on the calculation of the pairwise variation (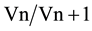 ) observed after the sequential addition of reference genes. A cut-off of 0.15 is typically used as a value below which there is no benefit of including other reference genes [22] . V values below the 0.15 threshold in Table 2 for the V2/3 combination show that proper normalization of gene expression in all our alfalfa sample sets could be achieved with only two reference genes. In accordance with our observations, Chen et al. [12] also observed that two stable reference genes were sufficient for accurate normalization of gene expression in banana (Musa acuminate) fruits. Their results also concur with our observation in Figure 1(b) that the combination of reference genes does however vary with experimental conditions.
) observed after the sequential addition of reference genes. A cut-off of 0.15 is typically used as a value below which there is no benefit of including other reference genes [22] . V values below the 0.15 threshold in Table 2 for the V2/3 combination show that proper normalization of gene expression in all our alfalfa sample sets could be achieved with only two reference genes. In accordance with our observations, Chen et al. [12] also observed that two stable reference genes were sufficient for accurate normalization of gene expression in banana (Musa acuminate) fruits. Their results also concur with our observation in Figure 1(b) that the combination of reference genes does however vary with experimental conditions.
3.3. Validation of Reference Genes
RT-qPCR determinations of relative levels of expression of sucrose synthase (SuSy), sucrose phosphate synthase (SPS) and proline dehydrogenase (ProDh) known to be involved in the cold acclimation process highlight the importance of using validated reference genes (Figure 4). When the two most stable reference genes UBL- 2a and 18S rRNA in Figure 1(a) were used for normalization of the 2011 cold acclimation samples, the results indicated a significant drop in SuSy and ProDh transcripts and a slight but significant increase in SPS gene expression in plants acclimated to low temperature. These results concur with previous observations that the enzymatic activity of SPS and SuSy are respectively induced and supressed in cold-acclimated crowns of alfalfa [37] . They are also consistent with the fact that ProDh is considered as a key enzyme in proline catabolism and that its expression is generally suppressed in plants exposed to abiotic stresses [38] . The slight but significant difference in SuSy expression between acclimated plants of ATF0 and ATF5 is indicative of the sensitivity of RT-qPCR analysis with validated reference genes.
![]()
Table 2. Pairwise variation analysis of the optimal number of references genes estimated by the geNorm V value. The V value is an estimate of the benefits of adding additional reference genes to the normalization factor. A V value <0.15 indi- cates no significant gain from the inclusion of additional reference genes.
![]()
Figure 4. Relative quantification of the expression of sucrose synthase, sucrose phosphate synthase and proline dehydrogenase in plants of the population ATF5 and of the initial genetic background (ATF0) sampled during the 2011 cold acclimation experiment. Expression was normalized with validated reference genes (UBL-2a and 18S rRNA) or with commonly used reference genes that were above the 0.5 M value threshold. Each bar represents the mean of four replicates for non-acclimated (NA) and cold-acclimated (A) plants of populations ATF0 and ATF5. Within each panel, treatments with the same letter are not statistically different at P > 0.05.
Normalization with less stable Act and Tub genes led to strikingly different conclusions including a lack of a low temperature effect on SuSy expression, a much higher increase in SPS gene expression and unexpected cold inducibility of ProDh. Such discrepancies are the results of biased adjustment to the data set when low temperature-repressed Act and Tub (Figure 2) are used to normalize gene expression. Xia et al. [32] noted that gene expression data normalized with less stable reference genes can lead to different conclusions due to weaker variation or higher levels of expression of target genes than those obtained with candidates validated with statistical algorithms.
4. Conclusion
Comparative geNormPlus analysis of a repertoire of candidate reference genes identified a set of candidates with high potential as reference for normalization in RT-qPCR analysis of gene expression in alfalfa. Genes encoding ubiquitin protein ligase 2a (UBL-2a), actin depolymerizing factor (ADF), retention in the endoplasmic reticulum protein 1 (Rer1) and eukaryotic elongation factor 1-alpha (eEF-1α) were found to be good references to normalize the expression of genes potentially related to key traits in alfalfa populations. Standardization of transcript levels in samples from a wide range of experimental conditions was systematically achieved with a combination of no more than two reference genes. When RT-qPCR data were normalized with stably expressed genes, observations on the expression of genes functionally related to cold acclimation in alfalfa was consistent with current knowledge of the physiology of that trait. Normalization with unstable genes led to markedly different conclusions with regard to the expression of these genes under these treatments. The candidate reference genes identified in our study will foster the acquisition of accurate RT-qPCR results in functional analyses of gene expression in alfalfa.
Acknowledgements
The authors would like to thank Dr. Serge Laberge for providing access to the EST library from cold acclimated alfalfa. We would also like to thank Mr. Réjean Desgagné and Mr. David Gagné for their contribution to the development of the EST collection and database. The participation of Dr. Marie-Pier Dubé to this project was supported by a NSERC visiting fellowship in Canadian federal government laboratories. This project was supported by a competitive grant of Agriculture and Agri-Food Canada.