Characterization of an Early Berry Development Grapevine Somatic Variant (Vitis labrusca L. cv. Isabel Precoce) ()
1. Introduction
Grapevine produces a fleshy fruit that is initially developed from two carpels separated by a septum, which results from the fusion of both carpels from the central tissue. After fertilization, the grape berry develops in two growth phases, which last approximately 30 and 50 days, respectively, separated by a 10 - 20-day lag phase. The initial growth phase during the first 15 - 30 days is characterized by intense cell division and enlargement associated with tartaric and malic acid accumulation. The second growth phase results exclusively from cell enlargement associated with the accumulation of hexoses during ripening [1] . Grapes are non-climacteric fruits and given their economic importance, berry ripening has been extensively investigated in the past decade, especially after its genomic sequence was made publicly available [2] . In spite of the advances and efforts made in grape genetics and genomics, little is known about the regulation of its ripening-related genes [3] - [12] . At the molecular level, significant advances have recently been made on the identification of genes involved in the grapevine fruit and flower development [1] [13] - [17] . Boss et al. [18] studied a mutant derived from the cultivar Pinot Meunier, which produces flowers where tendrils are normally formed, and identified a gene homologous to the gibberellic acid insensitive (GAI) from Arabidopsis related to the phenotype. Kobayashi et al. [19] demonstrated that the insertion of a retrotransposon in the promoter region of the gene VvMYBA1 is responsible for the absence of colour in white grapes. Fernandez et al. [16] [20] described a “fleshless berry” (Flb) mutant and identified five genes: a specific tissue protein 2, ATH13, a BURP domain protein, PISTILLATA and YABBY2, as candidate genes potentially involved with the phenotype. More recently, the same authors demonstrated that an insertion of a miniature inverted-repeat transposable element in the promoter region of the PISTILLATA-like (VvPI) gene results in a mis-expression pattern of the VvPI gene, impairing proper fruit development in grapevine [21] . Chatelet et al. [16] also characterized somatic variants that display different abnormal grape flower formation and development; Reiterated Reproductive Meristems (RRM) shows flower clusters with exacerbated ramification causing a delay in flower meristem specification. The RRM mutant phenotype is a result from the VvTFL1A cis-activation caused by an insertion of a class-II transposon in its promoter region, enhancing its expression [22] . Multiple Perianth Whorls (MPW) is impaired in the specification of anther and carpel identity [16] . CLS (Carpel-less) Mouvèdre variant exhibited alterations in the innermost whorls with the development of carpelloid structures. In this analysis, VvMADS1, VvMADS2, VvMADS3, putative orthologues of Arabidopsis flowering genes AG, SEP and AGL13 respectively, were evaluated by reverse transcription-quantitative PCR and showed differential expression (induction or repression) between the somatic variants compared with wild-types. VvMADS1 was strongly reduced in those somatic variants [16] . A recent detailed review made by Torregrosa et al. [23] describes the sources and the possible mechanisms involved in the somatic variations that have been observed in grapevine and describes other interesting grapevine somatic variants.
The present study describes the characterization of aspects of fruit growth and development from an “Early Berry Development” grapevine mutant known as cv. Isabel Precoce (Precocious Isabel), in which the time of fruit development is 30 - 35 days shorter, when compared to the wild type. “Isabel Precoce” is commercially cultivated in wine-growing regions of Southern Brazil and has agronomic advantages over “Isabel”, such as shorter life cycle, reduced fungicide applications, and less exposure to adverse weather conditions that may compromise yield and quality of the harvest. It is shown here that “Isabel Precoce” cultivar shares the microsatellite profile of “Isabel”, indicating that it can be considered as an Essentially Derived Variety (EDV) from “Isabel”. In an attempt to better characterize the early berry development phenotype of “Isabel Precoce”, comparison of physical and biochemical aspects and gene expression profiles during fruit development were investigated. Transcriptional profiles of a range of genes from different functional classes that are involved in flower and fruit morphogenesis as well as in ripening metabolism were used (e.g. MIKCC-type MADS box genes, sugar transport and control of flavonoid biosynthetic pathway). The results suggest that “Isabel Precoce” develops smaller fruits in diameter and volume, despite of following similar development kinetics and that gene expression profiles in developing berries of “Isabel” and “Isabel Precoce” are remarkably similar. Nevertheless, some differences in gene expression of the two cultivars have been identified, with a higher induction of transcript accumulation being observed in “Isabel Precoce”.
2. Material and Methods
2.1. Plant Material
In order to compare the two cultivars, the sampling strategy was based on monitoring developmental stages following the Meier scale [24] or chronologically, when “Isabel” and its EDV “Isabel Precoce” show their marked differences. Individual plants grown under a parral trellis conduction and controlled management at Embrapa Uva e Vinho were used as biological replicates. “Isabel” and “Isabel Precoce” plants were sampled in three growing seasons as described below. In the growing season of 2008/2009, berry samples following the Meier scale at Stage 57 (inflorescences fully developed, flowers separated), Stage 71 (fruit set: young fruits beginning to swell, flower remains loose), Stage 75 (berries pea-sized, bunches start to hang) and Stage 81 (beginning of ripening: berries begin to brighten in colour―véraison) were harvested and promptly frozen in liquid nitrogen just after picking in the field. At stage 81, berry samples before (with no color developed) and after onset of ripening (beginning of color development) were collected separately. Samples at Stage 81 were made on different days, because “Isabel Precoce” begins ripening early than “Isabel” (Figure 1). Berries were also collected chronologically in the growing season of 2005/2006 at fruit set (FS), 10, 40 and 80 days after fruit set (DAFS), and in the growing season of 2007/2008 at fruit set (FS), 15, 30 and 90 DAFS. For sampling at 80/90 DAFS, “Isabel Precoce” was close to harvest, while “Isabel” was still at véraison. The three types of sampling were used for Real-Time PCR experiments.
![]()
Figure 1. “Isabel” (a) at beginning of ripening (véraison―stage 81) and “Isabel Precoce”; (b) ready to be harvested at the same date. Graphical representation of the differences between “Isabel” and its derived somatic variant “Isabel Precoce”; (c) during the 2004/2005 production season.
The vineyards received, on average, two applications of fungicide per month. The average weather data of the berry growing season, from August to February, was: 921 hours of sunlight, wind speed of 6.6 m/s, 75% air relative humidity, 52 days of precipitation, 588.8 mm of rainfall and air temperature ranging from 16.8˚C to 22.8˚C.
2.2. Physical and Biochemical Aspects of Berry Growth
Berries from three plants of each cultivar (“Isabel” and “Isabel Precoce”) were harvested for biochemical and molecular analysis at 15, 30 and 90 DAFS in the summer of 2007-2008. For physical analysis, berries were measured twice a week after fruit set until 90 DAFS in the 2007/2008 season. Ten berries from the middle of each of three bunches per plant were randomly selected, measured for diameter (mm) and volume (mm3). Total soluble solids (SST) were also measured using a Palette Atago Pr-101 (0% - 45%) refractometer. Anthocyanins were also quantified as described by the following protocol. Berry skins from samples at the beginning and at the end of maturation were used for phenolic compounds extraction in 4 volumes (weight/volume) of hydroal- coholic solution (12% ethanol, 5.5 g tartaric acid, pH 3.2) during 36 hours at 28% and 100 rpm in absence of light. The samples were centrifuged for 5 min at 10.600 xg and a 2 ml aliquot of the supernatant was filtered in a Millipore HVLP 0.22 µm filter. For all samples, 20 µl was used for the HPLC injection. The anthocyanin quantification was done by HPLC using a Shimadzu binary pump system model LC10-AD, an UV-VIS array diode detector model SPD-M10A and a Rheodyne injector. The set was controlled by CLASS LC10 software from the same manufacturer. A C18 reverse phase column was used as the stationary phase and the mobile phase was composed by a gradient of the solvents A (acidified water with 5% formic acid v/v) and B (acidified methanol with 5% formic acid v/v). The solvent flow was 0.8 ml/min. The proportion of solvent B was added linearly from 20% to 100% in 50 minutes. The anthocyanins were identified by comparing retention time and the order of elution using malvidin chloride 3,5-diglucoside (Fluka) as standard. The anthocyanin quantification was determined by comparing the peak areas and expressed as total area percentage to all peaks.
Berry diameter and volume data were modeled using the Gompertz growth function, commonly used to describe growth curves [25] -[27] . In order to allow a double sigmoid curve and use parameters of easy interpretation, two Gompertz functions were added and the result was reparameterized into the function given by the equation:
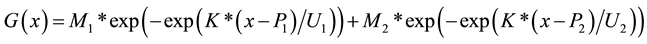
where:
M1, M2 = maximum size, in mm (diameter) or mm³ (volume) in each of the two phases (total maximum size = M1 + M2)
P1, P2 = precociousness, in number of days from fruit set to the two inflection points of the curve, where berry size is 36.8% of M1 or M2
U1, U2 = growth period, in days, from the moment when berry size is 10% of M1 or M2 until it reaches 90% of M1 or M2
K = −3.0844 (constant used to adjust U1 and U2)
x = time, in days after fruit set
M1, M2, P1, P2, U1 and U2 are the six parameters of the model, estimated using field data. Two separate curves were plotted, one for each cultivar. Because data collection ended before cv. Isabel reached vérasion, a simple sigmoid curve was used for that cultivar, by removing the second term of the function (setting M2 = 0, making P2 and U2 irrelevant). The significance of the difference between the two curves was asserted by testing this two-curve model with an F-test against an alternate model of a single curve combining both cultivars (see Supplementary Figure 1).
Statistical analysis was done using the R (2013) software, using non-linear model analysis, comparing curves with an F-test.
2.3. SSR-Based Genetic Identification Analysis
Total genomic DNA was extracted from young leaves of “Isabel” and “Isabel Precoce” grape cultivars using a modified procedure adapted from [28] [29] . The genetic identity between the original cultivar “Isabel” and its EDV “Isabel Precoce” was verified by genotyping nine microsatellite loci: VVMD36, VVMD27, VVMD8, VVMD7, VVMD7, VVMD6, VVS2, VVS4, VRZAG62, VRZAG72 as described by [30] . The estimated probability of identity between “Isabel” and “Isabel Precoce” was obtained as described by Paetkau et al. [31] using the Identity software [32] .
2.4. Selection of Candidate Genes for Real-Time PCR Analysis
In order to compare the normal “Isabel” and precocious “Isabel Precoce” phenotypes, the expression pattern of a range of genes from different functional classes that are potentially involved in flower and fruit morphogenesis as well as in ripening metabolism were screened from the literature and selected for real-time PCR analysis. Transcriptional profiles of the selected genes were then investigated during fruit development between both cultivars. The selected candidate genes and their correspondent functional classes are listed in Table 1.
![]()
![]()
![]()
Table 1. Selected genes evaluated between “Isabel” and “Isabel Precoce” fruit development and primers employed in (RT)- qPCR analysis.
aGene description based on classification according to GO terms and literature description. bDesigned for this work with Primer 3 Software (Rozen and Skalestsky, 2000).
2.5. Total RNA Isolation and Real-Time PCR Analysis
Total RNA was isolated from 100 - 150 mg of ground frozen tissues from pooled berries (whole berries, including skin and seeds) with Pure link® Plant RNA Reagent (Invitrogen), following the manufacturer’s protocol. The quality of RNA was verified by agarose gel electrophoresis and by the absorbance ratio A260/280 range from 1.8 to 2.0. Total RNA (0.5 μg) from all samples was treated with DNase I, followed by immediate double-strand cDNA synthesis using the SMART cDNA Synthesis Kit (Clontech) according to manufacturer’s instructions. Complementary DNA samples (10 μl, 100× diluted) from stages 57, 71 and 81 (2008/2009); 15, 30 and 90 DAFS (2007/2008) and 10, 40 and 80 DAFS (2005/2006) were added to each PCR reaction mix containing, 4 μM each primer, 1× SYBR Green, 50 µM dNTP, 1× PCR reaction buffer, 2 mM MgCl2 and 0.25 U Platinum Taq DNA Polimerase (Invitrogen) in a final volume of 20 μl. Thermocycling conditions were as follows: an initial enzyme activation of 5 min at 95˚C, followed by 40 cycles of denaturation for 15 s at 94˚C, anneling for 10 s at 60˚C, and extension for 15 s at 72˚C. The samples were kept at 40˚C for 120 s, followed by a melting point assay from 55˚C to 99˚C. The real-time PCR reactions were carried out in an Applied Biosystems StepOne Plus Real-Time PCR System. All templates were run in technical quadruplicates and three biological replicates. Sample values were normalized using the corresponding expression level of actin (VvACT), α-tubulin (Vv αTUBULIN) (data not shown) and expressed as the average standard deviation [33] . Specificity of the PCR product generated for each set of primers was tested by the melting gradient. The difference between the cycle threshold (Ct) of the target gene and the Ct of the constitutive genes established was used to obtain the normalized expression of the target genes, which corresponds to 2−ΔΔCt [34] . The statistical analyses were made by means of the Student t test with p < 0.05. The relative expression of each gene was gene-wise normalized using Genesis software [35] . Hierarchical clustering of gene expression data was performed using the Pearson correlation tool of the same software. Primer efficiencies were estimated according to Ramakers et al. [36] and Ruijter et al. [37] added to Table 1.
3. Results and Discussion
3.1. Description and Confirmation of Cultivar Clonal Identity
“Isabel Precoce” was identified in 1993 in a vineyard in Farroupilha, Rio Grande do Sul (Brazil; latitude: 29˚13′30″S, longitude: 51˚20′52″W) on a grapevine (Vitis labrusca L., cv Isabel) plant as a spontaneous somatic variant. The original plant was maintained in situ and cuttings were later grafted and propagated at Embrapa Uva e Vinho, in Bento Gonçalves (latitude: 29˚09′44″S, longitude: 51˚30′50″W), Rio Grande do Sul, Brazil, under the germplasm bank registration code 2526 [38] . “Isabel Precoce” plants were then followed for five production seasons to verify their phenotype, morphological and biochemical characteristics. As a result, the “Isabel Precoce” plants showed the same general phenotypic traits of “Isabel” and confirmed its early-ripening phenotypic stable feature, with an average harvest date of 33 days before its parental cultivar “Isabel” [38] . The growth cycle reduction occurs between flowering and harvest, particularly between flowering and the beginning of ripening (véraison―stage 81), when “Isabel Precoce” shows an accelerated early green berry development (see Figure 1(c)). “Isabel Precoce” was also cultivated in a semi-commercial scale in tropical areas of Brazil and demonstrated the same stable characteristics as in the south, where the climate is sub-tropical. Additionally, “Isabel Precoce” presents a remarkably even ripening of bunches when compared to “Isabel”, which ripens unevenly, with frequent occurrence of green berries in otherwise ripe bunches. The precociousness and uniform ripening of “Isabel Precoce” is greatly appreciated by farmers since the grapes can reach higher economic value.
Several genetically related cultivars are phenotypically similar and difficult to differentiate through morphological comparison [39] . In order to assist the identification process, Simple Sequence Repeat (SSR)-based molecular markers were used to confirm the identity of “Isabel Precoce”. The molecular profiles of the nine SSR loci from “Isabel” and “Isabel Precoce” were compared with the SSR profiles of the Vitis labrusca cultivars “Alba”, “Alexander”, “Champagne”, “Niagara Rosada”, “Vergennes”, “Concord” and “Concord Clone-30” from the germplasm bank at Embrapa Uva e Vinho (Table 2). The comparison of the SSR profiles showed that “Isabel Precoce” share the same molecular profile of “Isabel”, differing only by its precocious berry maturation phenotype. These results confirm previous data from the literature, which indicated that intra-varietal clones may differ considerably in phenotypes, in spite of displaying similar DNA profiles [30] [40] - [43] . The estimated probability of identity between “Isabel” and “Isabel Precoce” is 99.80%, therefore it is possible to conclude that “Isabel Precoce” is an EDV from “Isabel”.
![]()
Table 2. Microssatelite allele profiles of Vitis labrusca cultivars compared with “Isabel” and “Isabel Precoce”. Allele sizes are indicated in base pairs. nd = not determined.
3.2. Physical and Biochemical Changes during Fruit Development
In order to monitor fruit development differences between “Isabel” and “Isabel Precoce”, berries diameter and volume were measured. As shown in Figure 2, the Gompertz growth curves model berry growth adequately, both as diameter (R2 = 0.993) and volume (R2 = 0.988).
In the general comparison of the growth curves (both volume and diameter), a significant difference was detected (p < 0.0001) between both cultivars. Maximum diameter and volume estimated by the model were, respectively, 16.16 mm and 2498 mm3 for Isabel after the first growth phase, and 15.82 mm and 2096 mm3 for Isabel Precoce after both growth phases. It is expected that after the second growth phase, diameter and volume of Isabel will be even greater, emphasizing the smaller fruit size of Isabel Precoce. Maximum diameter and volume growth speed in the first phase were reached at 11.9 and 28.8 days after fruit set for Isabel, and 11.0 and 24.7 days for Isabel Precoce, respectively. Maximum growth speeds in the second phase of Isabel Precoce were reached at 56.8 days for diameter and 57.0 days for volume. The main growth period (10% to 90% of total growth) of diameter and volume of the first phase lasted 41.7 and 59.3 days for Isabel, and 44.0 and 36.0 days for Isabel Precoce, respectively. Although the diameter results apparently conflict with the volume results, graph analysis combined with the small difference between cultivars suggests that the lag phase between the two growth phases is larger in Isabel than in Isabel Precoce. The growth curves of Isabel Precoce are always similar or below the growth curves of Isabel, as seen in Figure 2. This suggests that the precociousness of “Isabel Precoce” is not related to accelerated berry growth. The main growth period of the second phase of Isabel Precoce lasted 5.0 days for diameter and 6.7 days for volume. The experiment ended with the harvest of Isabel Precoce, which occurred before vérasion in Isabel, therefore data for the second growth phase of Isabel is not available. Historic phenological data (Figure 1) has shown that, regardless of environmental conditions, the harvest point of Isabel Precoce has always been between 25 and 30 days before Isabel.
Usually, the development of grapevine berries is modeled based on a symmetric double sigmoid curve pattern [44] , with a phase of rapid growth during the first four weeks after flowering. A lag phase―characterized by a smaller number of cell divisions and an increase in cell size―takes place between four and eight weeks after flowering. This development pattern has been reported for other cultivars, such as “Cabernet Sauvignon” [45] . Even though the cultivars follow similar development kinetics, “Isabel Precoce” develops fruits of smaller diameter and volume (p < 0.0001) than “Isabel” after the first growth phase, and reaches the second growth phase about 42 days earlier (measured by the difference in vérasion). The results demonstrate that the Gompertz function can be used to accurately model berry development in grapevines.
![]()
Figure 2. Diameter ((a)―mm) and volume ((b)―mm3) variation during berry development of “Isabel” and “Isabel Precoce”. Data were collected twice a week after fruit set until 90 days after fruit set when “Isabel Precoce” was ready to harvest. Beginning and end of vérasion are indicated by dotted lines for each cultivar.
The content of total soluble solids (TSS) was also monitored for both cultivars. The TSS for “Isabel Precoce” was around 17.5˚Brix at 90 days after fruit set (ready for harvest). For “Isabel”, the content of total soluble solids at 90 DAFS―33 days before harvest―was around 14.9˚Brix. This difference in the TSS at the same time for both cultivars indicates that an accelerated biochemical process takes place earlier in “Isabel Precoce” when compared with “Isabel”. This parameter indicates a physiological difference between both cultivars, suggesting that precocity may be more closely related to changes in metabolic and biochemical characteristics. However, this difference is not clearly reflected in the diameter and volume parameters, as seen in Figure 2, which shows approximately the same development cycle for both cultivars. In both time scale (DAFS) and development scale [24] sampling, the same pattern of early berry development of “Isabel Precoce” was observed in biochemical and metabolic characteristics and also in gene expression profiles.
Anthocyanin content was measured using HPLC for both cultivars (Supplementary Figure 2). “Isabel Precoce” berry skin contains, on average, ten times more anthocyanins (A520 = 473.252 mg/ml fresh weight) than Isabel (45.304 mg/ml fresh weight) at 90 DAFS, confirming that the early anthocyanin accumulation marks the beginning of véraison in colored cultivars. Total anthocyanin accumulation were similar to other colored vinifera-type cultivars [20] , with Malvidin being the preferential predominant form. Acylated forms were predominant in the berry skin extracts from both cultivars. At 90 DAFS, when “Isabel Precoce” is ready to be harvested and “Isabel” is at véraison, “Isabel Precoce” had a free anthocyanin accumulation average of 57 mg/ml and 416 mg/ml of acylated anthocyanins. Isabel accumulated an average of 11 mg/ml of free anthocyanins and 69 mg/ml of acylated anthocyanins. Malvidin was the predominant free anthocyanin in “Isabel Precoce” extract, representing about 75% of total anthocyanins, whereas in “Isabel”, the most abundant free anthocyanin was cyanidin (36.8%).
3.3. Expression Profiles of Candidate Genes by Real-Time PCR
Analyses of gene expression under field conditions represents a challenge, since variation in environmental conditions can influence gene expression and ultimately grape berry ripening [46] - [49] . To compare and partially avoid environmental effects, two sampling strategies were used. The first was based on monitoring developmental stages and the second was time dependent. Individual plants grown under a parral trellis conduction and managed with controlled fertilization were used as biological replicates. The same plants were sampled as previously described in the materials and methods section.
In order to compare the ripening process of “Isabel” and “Isabel Precoce”, the expression patterns of a range of genes from different functional classes selected from the literature (listed in Table 1) were evaluated. The selection of the candidate genes followed a strategy based on gene description and its known association with distinct berry ripening phases and processes. Thus, since development of the berry skin color is the most remarkable phenotypic difference observed, three genes (VvMYBPA1, VvCHS and VvPAL) belonging to the phenylpropanoid metabolic pathway were selected (see Table 1). In an attempt to cover all the ripening phases, genes associated with flesh morphogenesis, cell expansion and known to be upregulated at early or late berry ripening stages were also selected, totalizing twenty five genes.
Initially, the gene expression analyses were performed using time-dependent sampling points collected during two different growing cycles (2005/2006 and 2007/2008 seasons), using a set of sixteen genes as shown in Figure 3. Genotypic and environmental effects were analyzed in order to detect differences in expression patterns between growing seasons and cultivars.
Hierarchical clustering analysis of gene expression patterns allowed the identification of two major clusters of transcription profiles that corresponded to genes preferentially expressed during in early and late stages of the berry development. At late ripening stages, the first cluster groups eight genes (VvExpansin, VvGRIP4, VvERE, VvADH2, VvCHS, VvMADS1, VvYAB2 and VvHT) with conserved expression patterns in both seasons and between cultivars. This cluster of genes presented the expected expression pattern as described in the literature referred on Table 1, being upregulated in the late stage of berry ripening, where important ripening metabolic processes (VvERE, VvCHS, VvADH2 and VvHT) and rapid cell expansion (VvExpansin, VvYAB2, VvMADS1, VvGRIP4) takes place.
The second mostly conserved expression group includes the eight genes that were downregulated at late stages of berry ripening (VvFT, VvMSA, VvHB13, VvSP2, VvBURP1, VvPAL, VvF3H and VvMYBPA1), but are more expressed at early green fruit development stages, where Isabel Precoce shows its accelerated green berry
![]()
Figure 3. Hierarchical clustering analysis of gene expression in “Isabel” and “Isabel Precoce” during fruit development for selected candidate genes (see Table 2 for details). Average intensity values are represented by a color scale for samples corresponding to fruit set (FS), 15, 30 and 90 DAFS for the growing season of 2005/2006 and fruit set (FS), 10, 40 and 80 DAFS for the growing season of 2007/2008. Expression analyses were performed by real-time PCR and relative gene expression data were normalized with VvActin and VvGAPDH. White points indicate the sample with absolute maximum expression for a gene measured in the experiment. Stars indicate statistical difference (p ≤ 0.05) between cultivars for the correspondent sample. Grey boxes indicate absence of gene expression.
development (Figure 1(c)). The observed transcription profiles for these genes are in accordance to the literature description, as referred in Table 1. The maximum absolute level of transcription was generally observed in Isabel Precoce samples, especially at the late stages of berry ripening, in agreement with the phenotypic characterization (Figure 1(c) and Figure 3).
Genome-wide expression analyses of MIKCC-type MADS Box genes during grapevine development suggested the recruitment of specific genes to regulate the development of specific grapevine organs, such as tendrils and berry fruits [50] . Therefore, gene expression profiles of seven MIKCC-type subfamily members were investigated: VvBS, VvSEP, VvAG, VvAGL15, VvAGL17.1, VvPI and VvAP3. In total, nine genes were evaluated in five representative stages of berry development during the growing season of 2008/2009. These genes were chosen because they are related to the establishment of the identity of floral meristems and were also identified during fruit development without a defined function [50] . For this analysis, VvMYBPA1 and VvExpansin genes were used as internal references, since their expression profiles are known to be upregulated at early and late stages of fruit development, respectively [4] [13] (and Figure 3). As shown in Figure 4, cluster analysis of gene expression profiles identified a main cluster of seven genes (VvBS1, VvSEP3, VvAG2, VvAG3, VvAGL15.1, VvAGL15.2, VvAGL17.1 and VvPI) that showed the highest expression levels at the pre-flowering (PF) stage for both cultivars, with a relevant downregulation in the following stages (FS, GB, PV and V), while VvBS2 presented a reverse kinetics. VvAG3 showed a distinct transcription profile between the cultivars, being significantly upregulated at pea-sized green berries (GB) in “Isabel Precoce” when compared to “Isabel”. Except for VvPI, VvAG2, VvAG3 and VvAGL15.2, the gene expression profiles observed for VvAGL15.1, VvBS1, VvBS2, VvSEP3 and VvAGL17.1 in our experiments followed an inverse kinetics across the developmental stages analyzed, when compared to the data reported by Riaz-Riquelme et al. [50] . This may be the result of inter- specific variation between V. labrusca (“Isabel” and “Isabel Precoce”) and the V. vinifera cultivar cv. Tempranillo. The VvAG3 contrasting transcription profiles observed between “Isabel” and “Isabel Precoce” (Figure 4), combined with literature information, prompted us to better investigate its possible association with the precocious phenotype from “Isabel Precoce”. The grapevine gene VvAG3 (also known as VvAGL11) is homologous to Arabidopsis gene AGL11 [18] that was shown to be expressed in developing ovules [51] . These genes are also similar to FBP7 and FBP11 from petunia, which have been shown to be involved in ovule and seed development [52] . Mejia et al. [53] proposed VvAG3 as a major gene to control seed development in grapes. More recently, Malabarba and co-workers (unpublished data) demonstrated that the absence of VvAG3 expression impairs the proper development of seed tissues. In grapevine, the first steps of berry development are under the control of hormones (auxins, cytokinins and gibberellins) that promote cell division and expansion [54] . Although these hormones can be imported into the berry, they are mostly produced by the seeds or maternal tissues
![]()
Figure 4. Hierarchical clustering analysis of gene expression in “Isabel” and “Isabel Precoce” during fruit development for selected candidate genes (see Table 2 for details). Average intensity values are represented by a color scale for samples corresponding to flowers (F), fruit set (FS), pea-sized green berries (GB), pre-véraison (PV) and véraison (V). Expression analyses were performed by real-time PCR and relative gene expression data were normalized with VvActin and VvGAPDH. The white dots denote the maximum average level of gene expression for the different development stages in the experiment. Stars indicate statistical difference (p≤ 0.05) between cultivars at same stage of fruit development. Grey boxes indicate absence of gene expression.
[54] . Therefore, it is tempting to speculate that the higher expression observed for VvAG3 gene in “Isabel Precoce” may help the seeds to follow a more uniform maturation process, which can result in synchronized and accelerated fruit development, mediated by smaller variations in hormonal production by the seed. This alternative interpretation is supported by the remarkable even ripening of the “Isabel Precoce” bunches, when compared to “Isabel”.
In the perspective of the data generated by this work, comparing physical and biochemical aspects and gene expression profiles during fruit development between “Isabel” and its derived mutant “Isabel Precoce”, we were able to describe an unique early berry development somatic variant in grapevine. In summary, the results suggest that “Isabel Precoce” develops smaller fruits and that transcriptional profiles of selected candidate genes in developing berries of “Isabel” and “Isabel Precoce” are remarkably similar. However, some differences in gene expression between the two cultivars have been identified, with a general higher induction of transcript accumulation being observed in “Isabel Precoce” (Figure 3 and Figure 4). The data obtained here provide the initial clues to guide the generation of additional knowledge associated with the early berry development phenotype. We are currently performing a genetic analysis approach in order to evaluate the heritability of this trait and to narrow the search for genes and genomic regions associated with the precocious berry development.
Acknowledgements
We are grateful to M.Sc. Umberto Almeida Camargo for providing us information and data from “Isabel Precoce” which were used to guide our experiments. This work was supported by CAPES (Coordenação de Aper- feiçoamento de Pessoal de Nível Superior), CNPq (Conselho Nacional de Desenvolvimento Científico e Tecnológico Proc. 475440/2004-8), FAPERGS (Fundação de Amparo à Pesquisa do Estado do Rio Grande do Sul Proc. 04/1184.6) and Embrapa.
Supplementary
![]()
Figure 1. Graphical representations of the analysis of variance of berry diameter and berry volume. The thick solid line represents the variance due to the use of a general model for both cultivars. The equation on the left side and the R2 and probability above it represent and test the general (single- curve) model. The effect of splitting the general model into two curves, one for each cultivar, is represented on the right of the thick line, with the significance of the F-test and the separate equations for each cultivar. The R2 of a combined model with two curves is represented on the left, below the R2 of the single-curve model. The error variance is represented by the dotted lines on the right, and the corresponding coefficient of variance (CV) is shown on the top right. Variances are represented as fractions of R2 using the scale on the bottom (horizontal lines are proportional to variances).
![]()
Figure 2. Quantification of anthocyanins in Isabel and Isabel Precoce cultivars. Sampling performed at 80 DAFS during the 2008/2009 season.
NOTES
*Corresponding author.