Optimization of Parameters for the Production of Lipase from Pseudomonas sp. BUP6 by Solid State Fermentation ()
1. Introduction
Lipases (triacylglycerol acyl hydrolase EC 3.1.1.3) are a cluster of enzymes catalyzing the hydrolysis of triacyl- glycerol to form glycerol and free fatty acids. In contrast to esterase, lipases are activated only at an oil-water interface. Many lipases are catalyzing a number of useful reactions including hydrolysis, esterification, trances- terification, acidolysis, alcholysis, and synthesis of peptides [1] . The extensive utilization of lipase has wide range of applications in synthesis of detergents, biosurfactants; organic, oleo-chemical, leather, cosmetic, per- fume, dairy and agrochemical industries; environmental management; biosensors, etc.
Microorganisms with potentials for producing lipases can be found in different habitats, including wastes of vegetable oils and dairy industries, soils contaminated with oils, seeds, and deteriorated food [2] . Since, lipase is among the most widely used class of enzymes in biotechnological applications and organic chemistry [3] , utili- zation of agro-industrial wastes as alternative sources of substrates would help solving pollution problems. The nature of the substrate is the most important factor affecting fermentative processes. The choice of the substrate depends upon several factors; mainly related to cost and availability.
Lipases are produced by several microorganisms, viz., bacteria, fungi, yeast, actinomycetes, archea, eucarya, etc. Microbial genera involved in the commercial production of lipases include: Candida, Mucor, Rhizopus, As- pergillus, Penicillium, Geotrichum, Rhizomucor, Bacillus, Pseudomonas and Staphylococcus [4] . Since they perform at wide range of pH and temperature, and they can be produced by solid-state fermentation (SSF) as well as submerged fermentation (SmF), microbial lipases are considered as highly robust. By dint of low pro- duction cost, greater stability, more simplicity and wider availability than SmF, SSF is considered as a novel strategy with higher physiological significance and industrial potentials [5] . Lipases are inducible enzymes; hence, different natural agro-industrial residues such as brans of wheat and rice, sugar cane bagasse, wastes from vegetable oil-refining, etc. can effectively be used as inducers or substrates for lipase production employing SSF strategy [4] -[8] . Deoiled kernels (i.e., cakes) from coconut, olive, gingelly, cotton, Jatropha, etc., obtained after extracting oil have been utilized as solid substrate for the fermentative production of lipases and other industrial enzymes. This is because residual oil and other ingredients contained in it serve as inducers for lipase production [15] .
Optimization of environmental parameters as well as the culture parameters may enhance the production of value-added products of commercial interest to many folds. Usually, “one parameter at a time” strategy was used for the optimization; but, this method is time consuming and hectic, and that the utilization of statistical tools makes the process easier. Response surface methodology (RSM) is such a kind of statistical tool being ap- plied widely for the optimization, modeling and analysis of problems related to the production of biomolecules [6] . However, the use of different substrates as well as cultivation strategies for the production of lipase still re- mains an emerging area of research. In this context, the present study focused on 1) screening of different oil cakes as solid substrate for the production of lipase by a novel rumen bacterium Pseudomonas sp. strain BUP6, and 2) statistical optimization of parameters for lipase production employing RSM technique.
2. Materials and Methods
2.1. Materials
Analytical and bacteriological-grade chemicals from Himedia (India) and Merck India Ltd. were used for the present study.
2.2. Culture Medium
The pure bacterial culture Pseudomonas sp. strain BUP6 (Genbank Accession No. KF 550910), isolated from the rumen of Malabari goat, was used for this study [9] . The bacterium was cultured on basal salt medium (BSM), supplemented with 0.3% of vegetable oil, and incubated at 37˚C for 24 h. The stock culture was main- tained on BSM agar slants, which was sub-cultured in an interval of 2 weeks. The BSM contained the following ingredients (%): 0.5 NH4NO3; 0.4(NH4)2SO4; 0.3 yeast extract; 0.2 K2HPO4; 0.2 NaCl; 0.01 MgSO4. 7H2O and 0.01 CaCl2 in double distilled water.
2.3. SSF Using Agro-Industrial Residues
Deoiled cakes of groundnut (GNC), gingelly (GOC), coconut (COC), soybean (SOC), and cotton seed (CSC) procured from the local market were used as solid substrate-cum-inducer for the production of lipase by SSF. Fermentation was carried out in 100 mL of conical flasks. Five grams of substrate (oil cake) were transferred into 100 mL conical flasks, and then moistened with10 mL of BSM (50%). In order to check the effect of pH on lipase production, the initial pH of the medium was set at 5, 7 and 9. All preparations in the flask were autoc- laved at 121˚C for 15 min, and inoculated with 0.1 mL of inoculum under aseptic condition. Production of lipase was assayed at regular intervals of 24 h for 5 days.
2.4. Extraction of Crude Enzyme
After incubation, the fermented matter in the whole flask was used for lipase assay at regular intervals of 24 h; for the extraction of crude lipase, 10 mL of 0.1 M Tris-HCl buffer was added to the flask and stirred for 10 min. Then the contents of the flask were centrifuged at 9400× g for 15 min at 4˚C, the supernatant was used as crude lipase for the activity assay.
2.5. Lipase Activity Assay
Production of lipase was qualitatively assayed by the method of by Priji et al. [9] . Para-nitro phenyl palmitate (p-NPP) was used as substrate for lipase assay. The assay mixture containing 1.8 ml of 0.1 M Tris-HCl buffer, 0.15 M NaCl and 0.5% Triton X-100 was pre-incubated with 200 µl of cell-free culture supernatant at 37˚C for 10 min. Subsequently, 20 µl of substrate (50 mM p-NPP in acetonitrile) was added to the reaction mixture and incubated at 37˚C for 30 min. The quantity of p-NP liberated was measured spectrophotometrically at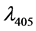 . One unit of lipase corresponds to 1 µmol of p-NP liberated per minute under the standard assay conditions. The li- pase activity was calculated as following Equation (1).
. One unit of lipase corresponds to 1 µmol of p-NP liberated per minute under the standard assay conditions. The li- pase activity was calculated as following Equation (1).
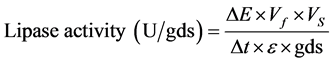 (1)
(1)
where —absorbance at 405 nm;
—absorbance at 405 nm;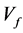 —final volume;
—final volume;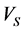 —volume of lipase used;
—volume of lipase used;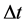 —time of hydrolysis;
—time of hydrolysis;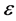 —extinction co-efficient (0.017); gds—dry weight in grams.
—extinction co-efficient (0.017); gds—dry weight in grams.
3. Statistical Optimization of Lipase Production
Box-Behnken Design (BBD), followed by RSM was employed to develop a mathematical correlation between different independent variables such as temperature, pH, moisture, and incubation time on the production of li- pase. The software Minitab version 14 (Minitab USA) was used to generate data and to analyze the experimental design of BBD and RSM.
3.1. Box-Behnken Design (BBD)
The culture parameters like temperature (28˚C to 40˚C), pH (5 to 9), moisture (20% to 60%), and incubation time (1 to 5 d) were selected for BBD. BBD at three levels (+1, 0, and −1)—designated as high, medium and low—was used for this study. Based on this design, a set of 27 experimental trials were suggested by the soft- ware. All the trials were carried out in duplicates and the results were analyzed by fitting to a second-order po- lynomial Equation (2). Each experimental trial was set up and lipase was harvested at proper intervals to meas- ure lipase activity as per the design.
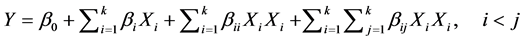 (2)
(2)
where 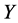 represents the response variable;
represents the response variable; 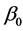 is the interception coefficient;
is the interception coefficient; 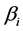 is the coefficient of the linear effect;
is the coefficient of the linear effect; 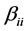 is the coefficient of quadratic effect;
is the coefficient of quadratic effect; 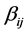 is the coefficient of interaction effect when
is the coefficient of interaction effect when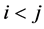 ; and
; and 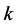 is the numbers of involved variables.
is the numbers of involved variables.
3.2. Validation Experiment
To check the validity of quadratic model, 4 experiments as predicted by point prediction software Minitab 14 were performed. Lipase activity was estimated and compared with predicted values.
4. Results
4.1. Effects of Different Substrates on Lipase Activity
Five different oil seed cakes (GNC, GOC, COC, SOC and CSC) were used as solid substrate-cum-inducer for the production of lipase by Pseudomonas sp. strain BUP6. GNC, GOC and COC supported the maximum pro- duction of lipase (107.44 U/gds, 86.21 U/gds and 9.97 U/gds, respectively) on Day 3 of incubation, subsequently the lipase activity was decreased sharply; while SOC as well as CSC supported the maximum production of lipase after 24 h of incubation, but in lesser amounts (95.74 U/gds and 5.45 U/gds, respectively) (Figure 1). Of them, GOC supported the highest lipase production (107.44 U/gds) (Figure 1). Effects of SOC (95.74 U/gds) and GOC (86.21 U/gds) were comparable to that of GNC (107.44 U/gds), but the other two oil cakes (COC and CSC) showed much lesser activity, i.e., 9.97 and 5.45 U/gds (Table 1).
![]()
Figure 1. Liapse production by Pseudomonas sp. strain BUP6 on different oil cakes.
![]()
Table 1. Maximum lipase activity in different substrates.
4.2. Statistical Optimization of Lipase Production
GNC which supported the maximum production of lipase was selected as substrate for the optimization studies. Four parameters (temperature, pH, moisture, and incubation) were considered for BBD analysis, followed by RSM to find out the optimum conditions for maximizing the production of lipase. A set of 27 experiments was conducted according to BBD, and the results showed that the predicted and experimental values for lipase activ- ities did not show significant difference (Table 2), i.e., the R2 value was 0.95-close to unity. A second order po- lynomial equation was fitted to the experimental lipase activity, which resulted in the following regression Equ- ation (3).
![]() (3)
(3)
where![]() —pH,
—pH,![]() —Temperature,
—Temperature,![]() —Moisture,
—Moisture,![]() —Incubation.
—Incubation.
The results were analyzed by standard analysis of ANOVA (Table 3). Based on these results, the model was utilized to generate response surfaces for the analysis of the variable effect on the production of lipase. The re- sponse surface plots obtained using Equation (3) is depicted in Figure 2.
![]()
Table 2. Experimental trials according to BBD model for the optimization of lipase production.
![]()
Table 3. Analysis of variance for the second-order polynomial model for optimization of lipase production [Degree of free- dom (DF), Sequential sum of square (Seq SS), Adjacent sum of square (Adj SS), Adjacent mean square (Adj MS), Text of statistics (F), Probability (P)].
4.3. Validation of Lipase Production
Four random experimental conditions were evaluated for the validation of the model. In all these instances, model prediction was in good agreement with the experimental data (considering the experimental error), and correlation coefficient was found to be 0.96 (Table 4).
Correlation coefficient was close to 1.0, suggesting the significance of the model. The optimum production of lipase was found to be 180.75 U/gds (at 28˚C, pH 5.9, moisture 33%, and incubation period 2 d). Thus, the sta- tistical optimization resulted in 0.7 fold of lipase activity over the unoptimized condition.
5. Discussion
Research on lipase progressed very rapidly during the past few decades, giving much emphasis on the exploita- tion and recycling of agro-industrial residues. The present study proved that Psuedomonas sp. strain BUP6 is an efficient producer of lipase on solid medium (oil cakes). Nowadays, SSF strategy is increasingly employed as a method for the production of lipase on different lipid-bound waste materials, because of the several advantages of SSF such as better yield and easy to control. As a part of this study, different natural substrate such as COC, GNC, SOC, GOC, and CSC were screened to spot out the best solid medium for lipase production. Of them, GNC supported the highest production of lipase (107.44 U/gds at 72 h). The cultivation period varies with the microorganism, i.e., fast growing bacteria were found to secrete lipase within 24 h [10] [11] . Bacteria normally grow in a complex nutrient medium containing carbon (oil, sugars, mixed carbon sources), nitrogen and phos- phorous sources and mineral salts. Other significant factors influencing lipase production include: initial pH, growth temperature, incubation period and moisture percentage (water activity). Temperature of the substrate during SSF critically affects the growth of microorganisms, and product formation [5] . Lipase yield by Pseudo- monas sp. BUP6 appeared to be dependent on moisture content. Results showed that lipase production (ex- pressed as enzyme activity) gradually increased from 20% to 60% moisture content and reached its maximum at 60%, and found that the moisture content was directly proportional to production of lipase; but high moisture content led to more contamination (may be due to poor aeration). The maximum lipase activity (114.75 U/gds) was obtained at 60% moisture content. Like moisture content, incubation time was also an important parameter that influenced the production of lipase. In the present study, 48 h of incubation was found to be optimum for lipase production. Incubation periods ranging from few hours to several days have been found to be best suited for the maximum lipase production by bacteria. For instance, an incubation period of 12 h was found optimum for the lipase production by Acinetobacter calcoaceticus and Bacillus sp. RSJ1 [10] and 16 h for B. thermoca- tenulatus [11] ; while, in the case of Pseudomonas sp., P. fragia and P. fluorescens BW 96CC, the maximum li- pase activity was obtained at 72 h and 96 h of incubation, respectively [12] [13] . Employing Candida rugosa, Benjamin and Pandey [1] reported the use of mixed-solid substrate containing wheat bran and coconut oil cake for lipase production. Fermentation was carried out for 72 h, and the maximum lipase yield was 118.2 U/g. Si- milarly, in the present study, the maximum activity of lipase obtained (107.44 U/gds) was at 72 h of incubation using GNC as substrate. It seems that the residual oil in the cake acted as both inducer and additional nutrient.
![]()
Figure 2. Response surface plots (3D) showing the effects of different parameters (X1: pH; X2: temperature, ˚C; X3: moisture, %; and X4: incubation time, d) on production of lipase by Pseudomonas sp. strain BUP6.
![]()
Table 4. Experimental trails for the validation of the predicted model.
Therefore, at the later stage of fermentation, the lipase activity was decreased; this might be an indication of the depletion of nutrient in the medium, i.e., lipase production is dependent on the nutrient status.
Few studies reported that oil cakes as the best solid substrate-cum-inducer for the production of lipase by SSF [14] [15] . D’Annibale et al. [16] reported that olive mill waste water as a growth medium for lipase production, which showed the highest lipase activity of 9.23 U/ml. Brozzoli et al. [17] studied the lipase production in bench-top reactor using the olive mill waste water medium and obtained the maximum production as 20.4 U/ml. Salihu et al. [18] used the statistical optimization of nutrient components to enhance lipase production by C. cy- lindracea, and the maximum activity was 20.26 U/ml. Vishnupriya et al. [19] assessed the lipase production by Sterptomyces grisesus, and found the maximum activity as 51.9 U/ml. Compared to all these studies, present study reports higher lipase activity (180.75 U/gds) at optimized condition.
Various physico-chemical parameters evaluated for the maximum production of lipase. Four parameters like temperature, pH, moisture, and incubation time for the lipase production were statistically optimized. Optimiza- tion is a complex process, which can be performed in two different ways: conventional and modern methods. The conventional method of optimization defined as the one-at-time strategy, but modern multivariate RSM technique enables optimization of more than one parameter at a time [6] , which can be performed for assessing the relationship between environmental and cultural parameters, so as to produce 3D contour and surface plots. This is more effective than conventional methodology, i.e., effective, easier, faster and more economical [6] . Groundnut cake which supported the maximum lipase production was selected for the optimization process. Under optimized conditions (28˚C, pH 5.9, moisture 33%, and incubation 2 d), the maximum production of li- pase was 180.75 U/gds on GNC, which was 0.7 fold higher than that of unoptimized conditions. R2 value (0.95) represents the good fixity of experiments with predicted values.
6. Conclusion
Briefly, this study investigated the lipase activities of Pseudomonas sp. BUP6 on agro-industrial residues as substrate, which showed that groundnut oil cake was the best for enhancing lipase production on solid medium. By statistically optimizing the culture parameters, the lipase production was further enhanced. Thus, this study focuses on the need for the exploitation of agro-industrial residues for the production of industrially-significant and human-friendly biomolecules; moreover, its low cost increases the industrial potentials.
Acknowledgements
The financial assistance (Grant No. 026/SRSLS/2012/CSTE) from Kerala State Council for Science, Technology and Environment (KSCSTE), Government of Kerala, is gratefully acknowledged.
NOTES
*Corresponding author.