Feeding Strategies for Enrichment and Characterization of Anammox Biomass in a Sequencing Batch Reactor ()
1. Introduction
Nitrogen discharges in water bodies are a constant matter of concern due to the environmental and public health issues they create. Nitrogen compounds cause eutrophication, mainly due to the excessive growth of algal biomass, and ammonia is toxic to aquatic organisms. Moreover, ammonium is oxidized to nitrate, decreasing the amount of dissolved oxygen in aquatic ecosystems and causing mortality of the aerobic biota. Concerning human health, the ingestion of nitrate can cause irreversible conversion of hemoglobin to methemoglobin, leading to respiratory disorders. Therefore, the development of efficient technologies for nitrogen removal from effluents is of paramount importance.
Conventionally, the nitrification-denitrification process has been employed for nitrogen removal in wastewater treatment plants. However, the need for supplementing the wastewater with organic electron donors for denitrification, increased sludge yields and high aeration costs for producing nitrates are clear drawbacks that have been promoting the search for alternative and cost-effective technologies. In this context, the anammox (anaerobic ammonium oxidation) process has spurred interest for nitrogen removal, since ammonium can be oxidized by autotrophic microorganisms without the addition of molecular oxygen [1] . The anammox reaction involves ammonium oxidation using nitrite as electron acceptor and produces dinitrogen gas. Therefore, this process is extremely advantageous when it is coupled to short nitrification (nitritation). Compared to conventional nitrification/denitrification, the nitritation/anammox system saves 50% on required oxygen, 100% on the external car- bon source and 80% in biomass management [2] .
One of the main issues regarding the anammox process is obtaining biomass, since anammox bacteria cannot be cultivated by conventional microbiological techniques [3] . The phylogenetic 16S gene approach identified anammox bacteria as autotrophic and related to the Planctomycetes phylum. It was previously believed that the Planctomycetes had limited environmental relevance, but microbial ecology studies have shown that this bacteria group is ubiquitous [3] . In this way, more information on enrichment processes and phylogeny for anammox biomass must be acquired.
In the present study, anammox biomass was enriched from a nitrifying-denitrifying reactor sludge using different feed strategies in a Sequencing Batch Reactor (SBR), and the 16S gene approach (using both specific and universal primers) was used to assess the biomass phylogeny with regard to anammox bacteria and Bacteria domain. Additionally, specific anammox activity was measured on steady state reactor operation.
2. Material and Methods
2.1. Operation of the Sequencing Batch Reactor
The enrichment of anammox biomass was carried out in a SBR for its biomass retention, simplicity and homogeneity of mixture. The reactor consisted of a borosilicate glass with a diameter of 20.3 cm and a height of 17 cm. A six-bladed turbine carried out stirring at 50 rpm (each blade with length of 6.8 cm and height of 2 cm). The temperature was maintained at 37˚C ± 1˚C, which presented the highest anammox activity according to [4] . The total volume of SBR was 5 L, and 3 L was discarded from each batch. The reactor was inoculated with 2 L of sludge from an anaerobic-aerobic reactor employed in the treatment of an amino acid-producing industry (Ajinomoto, Pederneiras/SP, Brazil) [5] .
In the first 45 days of operation, pure argon (99.95%) was flushed in reactor bulking, to achieve anaerobiosis (20 mL/min). The effluent pH exhibited values of up to 9.1, which was higher than the optimal values observed for the anammox process [6] . Because of the high effluent pH, pure argon was replaced by an Ar/CO2 (97%/3%) mixture for pH control and as a supplemental carbon source from the 45th day of operation on. This Ar/CO2 mixture was continuously flushed into the reactor headspace, to prevent the entry of oxygen, from the 89th day of operation on.
Five feeding strategies were carried out (Table 1). Under strategy S1, filling step was set to 12 hours to avoid nitrite accumulation in the reactor, which can inhibit the anammox process, with a 56 h cycle (3-cicle/week). Under strategy S2, the reaction step was adjusted from 240 h (10 d) to 48 h (2 d), since the reactor was manually fed when it was observed total consumption of nitrite. Strategy S3 comprised a cycle of 168 h (one-cicle/week) with constant filling (except during settling and discharging), aiming for the growing of biomass, in this strategy the weekly Applied Nitrogen Load (ANL) was the similar to strategy S1. As the increase of biomass was not observed, feeding strategy (similar to S2) was again employed for total consumption of nitrite (strategy S4). Under strategy S5, the reactor was initially filled over 2.5 h, operating with a 24 h cycle and the influent 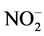 -N/
-N/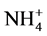 -N ratio changed to approximately stoichiometric requirements (Table 2).
-N ratio changed to approximately stoichiometric requirements (Table 2).
The SBR was fed with basal medium adapted from [7] . The ammonium and nitrite concentrations ranged
![]()
Table 1. Feeding strategies employed during the operation of the SBR.
1Total time of strategies.
![]()
Table 2. Ammonium and nitrite concentrations in each strategy.
1Ammonium and nitrite concentrations increased from 26 ± 4 to 65 mg 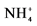 -N/L and from 23 ± 6 to 80 mg
-N/L and from 23 ± 6 to 80 mg 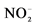 -N/L at last batch of strategy S2. 2Mean concentration in the reactor. Influent concentration were 204 mg
-N/L at last batch of strategy S2. 2Mean concentration in the reactor. Influent concentration were 204 mg 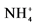 -N/L and 209 mg
-N/L and 209 mg 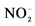 -N/L.
-N/L.
from 32 to 346 mg 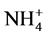 -N/L and from 24 to 456 mg
-N/L and from 24 to 456 mg 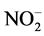 -N/L, respectively (Table 2). During the first 32 days of operation, 1.4 mg N2H4-N/L and 0.7 mg NH2OH-N/L were added for enhancing the enrichment of anammox biomass, since those compounds can restore the metabolic functions of anammox bacteria [8] . The medium was maintained in a glass vessel at 4˚C throughout the filling process.
-N/L, respectively (Table 2). During the first 32 days of operation, 1.4 mg N2H4-N/L and 0.7 mg NH2OH-N/L were added for enhancing the enrichment of anammox biomass, since those compounds can restore the metabolic functions of anammox bacteria [8] . The medium was maintained in a glass vessel at 4˚C throughout the filling process.
2.2. Analytical Procedures and Molecular Analyses
Ammonium, nitrate and nitrite were analyzed by flow injection analysis (FIA), in accordance with APHA [9] . The biomass concentration (as volatile suspended solids-VSS-concentration) was also measured according to APHA [9] . Specific anammox activity (SAA) was estimated from the slope of the curve describing the 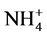 -N consumption as a function of time and related to the biomass concentration in 250 mL flasks filled with 90 mL of medium and 10 ml of anammox biomass.
-N consumption as a function of time and related to the biomass concentration in 250 mL flasks filled with 90 mL of medium and 10 ml of anammox biomass.
Biomass samples were collected for 16S rRNA gene analysis, at the end of strategy S3 (170th day of operation), which aimed for biomass increasing, and during strategy S5.4, which presented high anammox activity (450th day of operation). The DNA samples were amplified separately via PCR using anammox-specific (170th day of operation) and universal primers (450th day of operation), and the products were employed for the construction of 16S rDNA clone libraries. DNA segments consisting of approximately 780 base pairs (bp) of the 16S rRNA gene were PCR amplified using specific primers for anammox (46Frc-AMX820R) and the universal primers (6F/1510R) [10] according to the method described by [11] .
Extraction of total DNA was performed using the modified phenol-chloroform protocol described by [12] . The obtained PCR products were inserted into plasmids using the pGEM-T Easy Vector System. Competent DH5α E. coli cells were transformed with the plasmids (according to the manufacturer instructions) and clones were randomly selected. Nucleotide sequencing was performed in an ABI 310 PRISM DNA sequencer using the M13 forward primer. The obtained sequences were grouped and aligned using CLUSTALW [13] . After alignment, a distance matrix was created, which allowed the DOTUR software [14] to calculate the number of OTUs present at a 3% dissimilarity level. The sequences were compared with sequences in the NCBI database, to determine the phylogenetic identity of the clones, and a phylogenetic tree was created with the ARB software [15] , using the Lanemask PH filter and Thermothoga maritima as the outgroup.
The sequences of each OTU determined in this study were deposited in GenBank under accession numbers JQ609072 to JQ609091.
3. Results and Discussion
3.1. Operation of the Reactor
During the operation under strategy S1.1, no conversion of ammonium and nitrite was observed, since the pH values reached values up to 9.1, which is higher than the optimal values (7.0 - 8.5) [6] [16] . Despite the later pH decrease to 7.8 by sparging an Ar/CO2 (97%/3%) mixture, the nitrogen removal was still low (14% ± 6%) between the 45th and 55th day of operation. After 56 days of operation, the influent nitrogen concentration was reduced by half to avoid possible nitrite inhibition (S1.2). Under strategy S1.2, ammonium removal (37% ± 10%), but not nitrite removal, could be observed (Figure 1). The ammonium removal during strategy S1.2 was due to
![]()
Figure 1. Influent and effluent concentrations of nitrogen compounds under strategy S1 (A), S2 (B) and S5 (C). Ammonium influent (■), ammonium effluent (□), nitrite influent (●), nitrite effluent (○) and nitrate effluent(▲).
nitrification, which was verified by the presence of oxidized nitrogen compounds on the 89th day of operation (data not shown). Additionally, it was observed a biomass decrease at the end of strategy S1.1 (55th day of operation, Figure 2), probably due to the washing out of heterotrophic biomass. At this point, the reactor was filled for 12 hours, during which the ammonium concentration remained relatively constant, and nitrite levels increased until the 14th hour. Based on this observation, it was inferred that oxygen entered the reactor headspace during effluent withdrawal, allowing nitrification to occur until all of the dissolved oxygen had been consumed. Ammonium removal continued from this point, as 2.2 mg 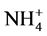 -N/L was observed to be removed until the end of this cycle without nitrification. For this reason, from this point on, a mixture of Ar/CO2 gases was continuously sparged into the reactor headspace to prevent the entry of oxygen.
-N/L was observed to be removed until the end of this cycle without nitrification. For this reason, from this point on, a mixture of Ar/CO2 gases was continuously sparged into the reactor headspace to prevent the entry of oxygen.
Under strategy S2, the SBR reaction time was determined by the observation of total consumption of nitrite (Figure 1(b)). Initially, the nitrogen compounds consuming time was 10 days. At the second batch of strategy S2, the reaction time decreased to six days, although a significant removal (total nitrogen removal rate of 60%) was obtained within 3 days. From the third batch on, the reaction time was established within two days, even when the concentrations of these compounds were increased punctually to approximately 80 mg 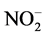 -N/L and 65 mg
-N/L and 65 mg 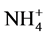 -N/L. The results obtained during strategy S2 indicated that nitrogen compounds concentration were adequate to obtain anammox activity after three months, similar to [17] and [18] . Moreover, the total nitrogen removal rate (60%) between the 118th and the 120th day of operation indicated a potential to remove higher nitrogen loads.
-N/L. The results obtained during strategy S2 indicated that nitrogen compounds concentration were adequate to obtain anammox activity after three months, similar to [17] and [18] . Moreover, the total nitrogen removal rate (60%) between the 118th and the 120th day of operation indicated a potential to remove higher nitrogen loads.
Under strategy S3 (121st to 179th day), the nitrite and ammonium concentrations were increased to approximately 210 mg of 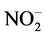 -N/L and 210 mg of NH4+-N/L, employing a 7-day cycle and constant reactor filling, except during settling and withdrawal (feed batch). The fed batch, in this case, was employed to maintain the same ANL as the one under the S1.1 strategy. Although almost 100% nitrogen removal was achieved, no increase of biomass was observed, since VSS concentrations in SBR were 0.66 ± 0.03 and 0.55 ± 0.18 g VSS/L on the 112nd and 177th days of operation, respectively. Probably, biomass increase was not observed because there was no increase of the ANL under strategy S3 (60 mg N/L·d).
-N/L and 210 mg of NH4+-N/L, employing a 7-day cycle and constant reactor filling, except during settling and withdrawal (feed batch). The fed batch, in this case, was employed to maintain the same ANL as the one under the S1.1 strategy. Although almost 100% nitrogen removal was achieved, no increase of biomass was observed, since VSS concentrations in SBR were 0.66 ± 0.03 and 0.55 ± 0.18 g VSS/L on the 112nd and 177th days of operation, respectively. Probably, biomass increase was not observed because there was no increase of the ANL under strategy S3 (60 mg N/L·d).
Since increase of biomass was not achieved during strategy S3, influent concentrations was returned to approximately 67 mg of ![]() -N/L and 64 mg
-N/L and 64 mg ![]() -N/L (strategy S4). Ammonium and nitrite removal were 66% ± 27% and 95% ± 9%, respectively, resulting in effluent concentrations of 2 ± 4 mg
-N/L (strategy S4). Ammonium and nitrite removal were 66% ± 27% and 95% ± 9%, respectively, resulting in effluent concentrations of 2 ± 4 mg ![]() -N/L and 18 ± 14 mg of
-N/L and 18 ± 14 mg of ![]() -N/L. The end of strategy S4 was considered to be the end of the biomass enrichment process, due to the high N removal efficiencies obtained.
-N/L. The end of strategy S4 was considered to be the end of the biomass enrichment process, due to the high N removal efficiencies obtained.
The main aspect that should be addressed at anammox biomass enrichment is the focus on maintaining anaerobiosis in the system including headspace. The main issues related to obtaining anammox activity under strategy S1 were related to allowing oxygen inside the SBR. Another important aspect to be considered is to gradually increase the ANL coupled with the gradually decrease of reaction time, according to the consumption of nitrite and ammonium.
At steady state reactor operation, five steps were adopted (strategy S5), by increasing the ANL up to 802 mg N/L·d, with total nitrogen removal efficiencies between 80% and 90% (Table 3). Under strategies S5.1, S5.2 and
![]()
Figure 2. Volatile Suspended Solids (VSS) concentration in the SBR.
![]()
Table 3. ANL, nitrogen load removed and total nitrogen removal during strategy S5.
S5.3, it could be seen that, as the ANL increased from 155 to 504 mg N/L·d, the SAA increased as well from 8.3 to 31.6 mg ![]() -N/g VSS·h (Table 4). The increase of the SAA up to strategy S5.3 was coupled to an increase of nitrite removed/ammonium removed ratio (N-Nitrite/N-Ammonium) and a decrease of nitrate produced/ ammonium removed ratio (N-Nitrate/N-Ammonium) (Figure 3). Employing a stoichiometric influent ratio may improve specific anammox activity [19] . Between strategies S5.1 and S5.3, the N-Nitrite/N-Ammonium ratio increased from 1.31 ± 0.12 to 1.40 ± 0.07 and the N-Nitrate/N-Ammonium ratio decreased from 0.29 ± 0.06 to 0.26 ± 0.05. At strategy S5.3, the N-Nitrite/N-Ammonium was higher than the theoretical ratio (1.32) and the N-Nitrate/N-Ammonium ratio approached the theoretical value (0.26) [20] . Moreover, the SAA after S5.1 increased 4‑fold at S5.3 suggesting that the proportion of the anammox bacterial population within the microbial consortium had increased (Table 4), despite the fact that the biomass concentration (VSS) remained stable (Figure 2).
-N/g VSS·h (Table 4). The increase of the SAA up to strategy S5.3 was coupled to an increase of nitrite removed/ammonium removed ratio (N-Nitrite/N-Ammonium) and a decrease of nitrate produced/ ammonium removed ratio (N-Nitrate/N-Ammonium) (Figure 3). Employing a stoichiometric influent ratio may improve specific anammox activity [19] . Between strategies S5.1 and S5.3, the N-Nitrite/N-Ammonium ratio increased from 1.31 ± 0.12 to 1.40 ± 0.07 and the N-Nitrate/N-Ammonium ratio decreased from 0.29 ± 0.06 to 0.26 ± 0.05. At strategy S5.3, the N-Nitrite/N-Ammonium was higher than the theoretical ratio (1.32) and the N-Nitrate/N-Ammonium ratio approached the theoretical value (0.26) [20] . Moreover, the SAA after S5.1 increased 4‑fold at S5.3 suggesting that the proportion of the anammox bacterial population within the microbial consortium had increased (Table 4), despite the fact that the biomass concentration (VSS) remained stable (Figure 2).
At strategy S5.4, the SAA was 29.4 mg ![]() -N/g VSS·h, which is similar to S5.3. On the 473th day of operation (strategy S5.5) the SAA was lower than the ones obtained in the S5.3 and S5.4, probably due to nitrite inhibition.
-N/g VSS·h, which is similar to S5.3. On the 473th day of operation (strategy S5.5) the SAA was lower than the ones obtained in the S5.3 and S5.4, probably due to nitrite inhibition.
Nitrite concentration on the second hour of cycle was 296.5 mg ![]() -N/L. Reference [21] observed inhibition at concentrations greater than 244 mg
-N/L. Reference [21] observed inhibition at concentrations greater than 244 mg ![]() -N/L, while [22] reported that 400 mg
-N/L, while [22] reported that 400 mg ![]() -N/L reduced in 50% the activity of granular biomass. Nevertheless, the SAA was 24.9 mg
-N/L reduced in 50% the activity of granular biomass. Nevertheless, the SAA was 24.9 mg ![]() -N/g VSS·h under strategy S5.5, even at higher nitrite influent concentrations (456 ± 14 mg
-N/g VSS·h under strategy S5.5, even at higher nitrite influent concentrations (456 ± 14 mg ![]() -N/L). The effluent nitrate concentrations were significantly lower than it would be expected, based on anammox metabolism, since it was observed a N-Nitrate/ N-Ammonium ratio of 0.19 ± 0.05. Because of the lower concentrations of nitrate in the effluent, it was observed the highest total nitrogen removal rate under strategy S5.5 (89.6%). Probably, nitrate-reducing activity was likely occurring during strategy S5.5. Anammox bacteria may have carried out this nitrate reduction: they exhibit a denitrifying pathway in which nitrate is reduced (using residual organic carbon) to ammonium via nitrite, which is then transformed into molecular nitrogen (N2) [23] .
-N/L). The effluent nitrate concentrations were significantly lower than it would be expected, based on anammox metabolism, since it was observed a N-Nitrate/ N-Ammonium ratio of 0.19 ± 0.05. Because of the lower concentrations of nitrate in the effluent, it was observed the highest total nitrogen removal rate under strategy S5.5 (89.6%). Probably, nitrate-reducing activity was likely occurring during strategy S5.5. Anammox bacteria may have carried out this nitrate reduction: they exhibit a denitrifying pathway in which nitrate is reduced (using residual organic carbon) to ammonium via nitrite, which is then transformed into molecular nitrogen (N2) [23] .
Therefore, nitrate may have been reduced to molecular nitrogen by the aforementioned pathway, since the effluent concentrations of ammonium were also reduced (i.e., nitrate generated via anammox activity was reduced to nitrite—by nitrate-reducing activity—and removed together with the remaining ammonium via anammox activity) (Figure 1(c)). Additionally, the observed nitrate reduction may also be due to other nitrate-reducing organisms (e.g., Pseudomonas sp. or other Proteobacteria species).
3.2. Bacterial Diversity
A total of 21 anammox-like 16S rDNA sequences were obtained after 170 days of operation. The sequences from 21 clones were grouped into two OTUs. OTU esp 1 (19 sequences) was 99% similar to Brocadia sinica and 92% similar to B. anammoxidans. OTU esp 2 (two sequences) showed 99% similarity to Anammoxoglobus propionicus. Figure 4 shows the phylogenetic tree containing the described OTUs.
The phylogenetic tree depicted in Figure 4 also contains the OTUs found in the SBR after 450 days of operation. The OTUs were divided into six major bacterial groups: Planctomycete, beta-Proteobacteria, green sulfur bacteria of the Chlorobi phylum, Nitrospira, Chloroflexi and the OP 11 candidate phylum. In a metagenomic study carried out by [24] , the following groups were found in anammox biomass: alpha-, beta-, gamma- and delta-Proteobacteria, Acidobacteria, Chloroflexi, Chlorobi, Bacteroidetes, Planctomycetes and the OP 11 candidate phylum.
![]()
Figure 3. Stoichiometric ratio of nitrite removed/ammonium removed (■) and nitrate produced/ammonium removed (▲) under strategy S5. Solid lines represent the theoretical molar ratio.
![]()
Figure 4. Phylogenetic tree of the examined 16S rRNA gene fragments based on OTUs esp 1 and 2 (●) and OTUs uni 1 - 19 (♦) found in samples from the SBR. Thermotoga maritima was used as the outgroup; the bar indicates one substitution per 100 nucleotides. The ARB program tree was constructed using a LanemaskPH filter.
![]()
Table 4. Applied nitrogen load (ANL), nitrogen load removed and total nitrogen removal during strategy S5.
It is likely that the OTU uni 12 - 14 clones, which contain sequences related to Pseudomonas and Comamonas, exhibited nitrate-reducing activity (as suggested by the results from S5.5). References [25] [26] showed that uncultured Nitrospira sp. could fix CO2 to produce pyruvate, but not acetate, butyrate or propionate in activated nitrifying sludge. Therefore, this unusual type of Nitrospira sp. metabolism indicates that short-chain organic acids may have acted as electron donors for nitrate-reducing microorganisms.
However, after 450 days of operation, Brocadia-like clones out-competed Anammoxoglobus propionicus. Reference [27] described a new species of anammox bacteria showing a different type of metabolism than had been previously described. They reported that A. propionicus could co-oxidize ammonium and propionic acid and presented a specific propionic acid removal rate of 0.64 mmol/g protein.min. Surprisingly, two anammox bacteria species were found in the SBR after 170 days of operation, but only the Brocadia species remained in the reactor after 450 days of operation. After long term period of SBR operation residual organic carbon of endogenous origin may not have been present, explaining the ability of Brocadia-like species to out-compete A. propionicus.
4. Conclusion
The results obtained in this study showed that sludge obtained from a pilot scale reactor treating amino-acid was a suitable inoculum source for enriching an anammox biomass. Maintaining anaerobiosis in the SBR and increasing ANL while decreasing reaction time were crucial procedures for successfully establishing the process. The SBR reached the conversion efficiency of 98.4% and nitrogen load removed of an average of 518.9 g N/m3∙day. A suitable approach to obtain a high biomass activity was to increase ANL coupled to an adequate influent ![]() -N/
-N/![]() -N ratio (near to 1.32 at S5.1-5). Moreover specific anammox activity was only partially inhibited by high nitrite concentration (296.5 mg
-N ratio (near to 1.32 at S5.1-5). Moreover specific anammox activity was only partially inhibited by high nitrite concentration (296.5 mg ![]() -N/L). After 170 days of operation, samples of the anammox biomass produced two OTUs: one was similar to B. sinica and the other was similar to Anammoxoglobus propionicus. Two species of anammox bacteria were found in the SBR with 170 days of operation, but only the Brocadia-like species remained in the SBR up to 470 days of operation, along with other species possibly related to the nitrogen cycle.
-N/L). After 170 days of operation, samples of the anammox biomass produced two OTUs: one was similar to B. sinica and the other was similar to Anammoxoglobus propionicus. Two species of anammox bacteria were found in the SBR with 170 days of operation, but only the Brocadia-like species remained in the SBR up to 470 days of operation, along with other species possibly related to the nitrogen cycle.
Acknowledgements
The authors gratefully acknowledge FAPESP (Fundação de Amparo à Pesquisa do Estado de São Paulo, Brazil) for financial support, CNPq (Conselho Nacional de Desenvolvimento Científico e Tecnológico, Brazil) for the grant, as well as Dr. Claudia Etchebehere and Dr. Dagoberto Yukio Okada for the careful reading of the manuscript.
NOTES
*Corresponding author.