1. Introduction
Phosphate is an essential inorganic molecule required for diverse cellular functions including nucleotide anabolism, bone remodeling and growth, and phosphorylation of a variety of substrates for function or augmentation of activities. Phosphate is required for life and thus a wide variety of mechanisms have evolved to acquire, store, and regulate phosphate metabolism.
The budding yeast Saccharomyces cerevisiae has served as an effective model organism for many cellular pathways, phosphate transport, accumulation, and storage is no exception [1] . Phosphate is actively transported across the plasma membrane via several phosphate transporters and is then transported into the vacuole (roughly equivalent to the mammalian lysosome but with additional metabolite storage functions) where it is formed into storage polyphosphate [2] [3] . Investigators have determined proteins required for polyphosphate accumulation and storage using DNA microarray technology [4] and mutant screening [5] . Polyphosphate accumulation in Saccharomyces cerevisiae is correlated to the growth phase [5] and the regulation of polyphosphate accumulation and breakdown has been analyzed [4] .
Phosphate storage in the form of the volutin granule has been well-characterized in a variety of microorganisms. Volutin was first identified in the early 1900s by its ability to be differentially stained by basic dyes in both bacterial cells and yeasts [6] . Later it was determined that these volutin granules contained polyphosphate in the form of linear polyphosphate chains of several to over 100 phosphates, covalently joined via high-energy phosphoanhydride bonds. Volutin is found not only in bacteria and yeasts, but also eukaryotes, such as trypanosomes, malaria parasites, Chlamydomonas reinhardtii, and Dictyostelium discoideum [6] . In certain eukaryotes, the volutin granule is a membrane-bound storage organelle for calcium as well as polyphosphate. This organelle was found to be acidic; this type of organelle is now called the acidocalcisome [6] .
Phosphate storage and accumulation in Saccharomyces cerevisiae is tightly controlled. Cells sense phosphate; and when the nutrient is low, transcription of many genes is upregulated through Pho4p, a transcription factor, and Pho2p, its regulator. Phosphate itself is transported into the cells via several transporters, including Pho89p, Pho84p, Pho87p, and Pho90p. When the intracellular phosphate level is high, the high affinity transporter Pho84p is targeted for degradation at the vacuole, and Pho4p is phosphorylated and excluded from the nucleus, and cells do not transcribe genes necessary for phosphate storage and accumulation [7] -[9] . When intracellular phosphate is low, Pho84p remains active; Pho4p binds to DNA and transcribes its target genes, and cells upregulate a myriad of genes necessary for phosphate storage and accumulation. When phosphate is low, several vacuolar proteins, Vtc1p, Vtc2p, Vtc3p, and Vtc4p, move from around the secretory membranes such as the endoplasmic reticulum to the vacuole [10] are necessary for polyphosphate formation [7] -[9] , with Vtc4p being the polyphosphate polymerase [11] . Polyphosphate in the vacuole has been well characterized biochemically in wild type and mutant strains under a wide variety of conditions [12] -[19] .
We have been studying volutin granule formation in wild type cells, in cell strains lacking the ability to traffic some or all proteins to the yeast vacuole (termed vacuolar protein sorting or vps mutants) [20] , and in cells treated with a variety of pharmacological agents. Intriguingly, several strains carrying a vps mutant are still able to make volutin and cells treated with inhibitors of vacuolar function are also able to form volutin.
2. Materials and Methods
2.1. Deletion Strain Analysis
All deletion analysis was performed in the parent strain BY4742 (MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0, obtained from the Yeast Genome Deletion Project) [21] or in deletion strains of BY4742 obtained from the Yeast Gene Deletion Collection (Invitrogen) with each gene, as indicated, replaced with KanMX. Cells were grown overnight at 30˚C in low phosphate YPD (made by precipitating out the majority of the phosphate using ammonium treatment followed by filtration, with an approximate concentration of phosphate of 0.01 to 0.03 mM) [22] and then were transferred into regular YPD media (1% yeast extract, 2% peptone, 2% dextrose, containing between 1 - 10 mM inorganic phosphate), and aliquots of cells were taken as indicated, harvested by centrifugation, stained for volutin [23] , and analyzed by microscopy with differential interference contrast optics. Volutin staining was accomplished by adding 100 μl volutin stain (4 ml aqueous toluidine blue, 20 ml formaldehyde and 0.6 ml glacial acetic acid) to the cells pellet [23] , vortexing, and allowing them to incubate for at least an hour before microscopic analysis. The t = 0 time point was taken before the media transfer. Pictures are representative of cells seen and correspond to experiments repeated at least three times. Magnification is 100X.
2.2. Quantitative Volutin Assessment
Wild type SEY6210 Saccharomyces cerevisiae (MATα his3Δ200 leu2-3, 112 lys2-801 trp1-Δ901 ura3-52 suc2- 3Δ9 GAL) [24] were grown overnight in low phosphate YPD [22] and then back diluted into low phosphate YPD to be grown for several more hours. A sample of cells was removed before any treatment and termed t = 0. It was centrifuged, the low phosphate media decanted and then volutin dye was added [23] . Cells grown in low phosphate media were centrifuged, transferred into high phosphate YPD (regular YPD) and were treated with the appropriate treatment. Ionomycin, cyclohexamide, concanamycin, and carbonyl cyanide m-chlorophenyl hydrazine (CCCP) were all obtained from Sigma and resuspended in DMSO and then added immediately at the indicated concentrations. Aloquits were removed at the indicated times of the time courses, cells were harvested by centrifugation and were stained for volutin [23] . Cell analysis was performed by light microscopy. The amount of volutin seen in the vacuole was scored: empty (none), low (up to 25%), medium (25% - 50%), or high (greater than 50%). Approximately 100 cells were counted per analysis. Statistical significance was performed by Chi-Square analysis.
3. Results and Discussion
3.1. Deletion Strain Analysis
We have been studying volutin granule formation in wild type cells and in cell strains lacking the ability to traffic some or all proteins to the yeast vacuole (termed vacuolar protein sorting or vps mutants). We have been able to induce volutin granule formation by growing the cells overnight in low phosphate media [22] and then switching the cells to high phosphate media [25] and staining with toluidine blue [23] . Volutin can be detected at the earliest time point we can stain (30 seconds) and continues to accumulate, reaching a peak after about 30 minutes and then slowly declining (data not shown). Volutin is not seen if cells grown in low phosphate media are reinoculated into low phosphate media, nor when cells are grown continuously in high phosphate media (data not shown). Volutin granules are seen exclusively in the vacuole of wild type cells (Figure 1). We were interested in determining the pathway by which volutin granule proteins are trafficked to the vacuole, and so undertook volutin granule staining in a series of vps mutants (Figures 2-6). Extensive volutin granule formation is seen in all mutant strains, except for Δvps33, a mutant cell strain lacking all vacuolar structure.
In Figures 1-6, cells were incubated in low phosphate media overnight at 30˚C, centrifuged and the low

Figure 2. Δvps21 cells. Vps21p is required for trafficking to the late endosome via both the early secretory pathway and also endocytosis [26] .

Figure 3. Δvps15. Vps15p regulates Vps34p, a PI3 kinase required for vacuolar protein sorting [27] .

Figure 4. Δvps27. Loss of VPS27 leads to formation of an exaggerated late endosome compartment and loss of traffic out of the late endosome [28] .

Figure 5. Δatg8. Atg8p is required for the formation of autophagosomal membranes and cytoplasm-to-vacuole targeting [29] -[31] .

Figure 6. Δvps33. Loss of VPS33 leads to a phenotype of total loss of vacuolar structure and function [32] .
phosphate media decanted. Dye was added and then the t = 0 time point was taken. For the remainder of the time points, high phosphate media was added, and then the cells were incubated at 30˚C for the indicated time periods. The cells were harvested by centrifugation, the media decanted, and volutin staining was performed [23] , and cells were viewed under differential interference contrast with a magnification of 100X.
Different blocks in vacuolar protein sorting seem to have differential effects on volutin granule accumulation. In vps33 cells, which lack all vacuolar structure, there is no granule formation (Figure 6). In vps21 cells (as well as vps9 [33] and vps45 [34] , data not shown), which have a block in protein sorting to the late endosome, granule formation is extensive (Figure 2). In cells lacking the protein serine-threonine kinase Vps15p, and thus the activity of the Vps34p PI3kinase, volutin accumulation is less than is seen in wild type (Figure 3), but is still markedly present. In vps27 cells blocked in their ability to traffic out of the prevacuolar compartment (PVC), volutin accumulation occurs, but to a lesser extent than wild type or vps21 cells (Figure 4). Due to the fact that there are large accumulations of volutin in the vacuole of vps21 mutant cells, we propose that proteins necessary for volutin accumulation are not trafficked through the secretory pathway via the endosome to the vacuole and must partake of a different route to the vacuole.
We had hypothesized this route is the cytosol-to-vacuole pathway (CVT) [31] . Proteins, such as aminopeptidase I (API), are trafficked directly from the cytosol to the vacuole in a membrane bound form (CVT) and organelles and cytosol are also transported to the vacuole for recycling under starvation conditions (autophagy) [29] [30] . We reasoned that perhaps low phosphate would trigger transport of proteins via autophagosome formation. Vps15p is required for API maturation in the CVT pathway and in a vps15 mutant all API sorting is blocked [35] ; and thus we would expect in the vps15 mutant the cytosol-to-vacuole pathway would be blocked and volutin would not accumulate. However, in Figure 3, in a vps15 mutant volutin still accumulates in the vacuole. Additionally, in Figure 5, volutin granules are still formed in an atg8 mutant, which is required for autophagy and cytoplasm-to-vacuole targeting [29] -[31] .
An alternative hypothesis is that the decrease in volutin formation is a pleiotropic effect due to an intrinsic physiological change in the vacuole, similar to what is seen when vacuoles are treated with an agent that raises vacuole pH [36] , mutations that perturb energy balance [5] , or vacuolar physiology [37] . This alternate hypothesis explains our data, as volutin granules are formed in all mutants tested.
3.2. Pharmacological Agent Analysis
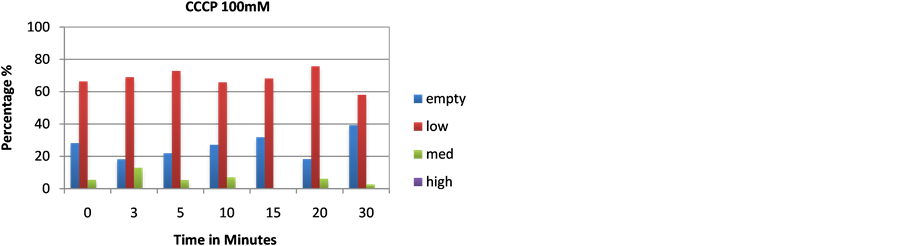
We were also interested in looking at the morphology of the granule after cells were treated with several pharmacological agents. There have been several recent reports of polyphosphate accumulation in yeast cells treated with agents targeting vacuolar function [13] [14] [17] [18] and these reports have implicated vacuolar physiology, such as the action of the ATPase. These reports analyze total polyphosphate accumulation biochemically. We sought to correlate cells treated with several agents with microscopic morphology of the volutin granule. Thus we undertook analysis of cells treated with cyclohexamide, carbonyl cyanide m-chlorophenyl hydrazine (CCCP), concanamycin A, or ionomycin, analyzing the formation of the volutin granule in the cell. Cells were scored as to the size of the dyed accumulation, as empty (none), low (5% - 25%), medium (25% - 50%), or high volutin (50%+) accumulation and graphed (Figures 7-11).
In order to determine if the treatments were statistically significant from each other, we performed Chi-Square analysis on each treatment end point (t = 30, final accumulation where we saw the highest accumulation percentages) comparing to the no treatment control. Only in the 1 μM concanamyacin A treatment there was no statistical difference between the treatment and control. In all the other treatments at the 30 minutes time point, Chi-Square analysis revealed a statistically significant difference (5 μg/ml cyclohexamide P < 0.001; 50 mM and 100 mM CCCP P < 0.001; 0.2 μM ionomycin p < 0.05; 0.4 μM ionomycin p < 0.001; 2 μM concanamycin A P< 0.05; 4 μM concanamycin A P < 0.001). Thus, all treatments decreased volutin accumulation over time.
In cells not treated with any agent, volutin staining increases over a 30-minute time course (Figure 7). However, when cells are treated with other agents, volutin accumulation decreases compared to the no treatment control. For example, in Figure 8, when cells are treated with cyclohexamide, a potent protein synthesis inhibitor,

Figure 7. Cells were transferred from low phosphate media to YPD and volutin accumulation was monitored microscopically.

Figure 8. Cells were transferred from low phosphate media to YPD with 5 μg/ml cyclohexamide and volutin accumulation was monitored microscopically.
 (a)
(a) (b)
(b) (c)
(c)
Figure 9. Cells were transferred from low phosphate media to YPD with concanamycin A ((a) 1 μM, (b) 2 μM, and (c) 4 μM) and volutin accumulation was monitored microscopically.
 (a)
(a) (b)
(b)
Figure 10. Cells were transferred from low phosphate media to YPD with CCCP ((a) 50 mM, (b) 100 mM) and volutin accumulation was monitored microscopically.
 (a)
(a) (b)
(b)
Figure 11. Cells were transferred from low phosphate media to YPD with ionomycin ((a) 0.2 μM, (b) 0.4 μM) and volutin accumulation was monitored microscopically.
less stained volutin is seen in the time course; however, there is still volutin made. This potentially indicates that protein synthesis plays a small role in generating protein players for polyphosphate accumulation after cells have been incubated for many hours in low phosphate medium. In cells treated with agents that directly inhibit vacuolar physiology, volutin staining is decreased as well. Concanamycin A is an inhibitor of the vacuolar ATPase; CCCP is a protonophore that will raise the pH of the vacuole of treated cells and obliterate the proton gradient; and ionomycin is an ionophore that allows calcium release from the vacuole. Each treatment decreased volutin staining over time in the treated cells. Each treatment changes one aspect of the vacuolar ion physiology. However, in each case volutin formation decreased.
We propose that the physiology of the vacuole itself, and the pH as well as the calcium content, are necessary for optimal volutin formation. In the case of the vps mutants small or large perturbations of the vacuolar milieu may be causing the volutin phenotype we see, rather than having a direct effect on the formation. Indeed, disturbing phosphate homeostasis leads to marked increases in calcium in Saccharomyces [38] . Future work should include analysis by additional assays [39] [40] and in many more vps strains, as well as additional pharmacological agents, such as wortmannin, an inhibitor of phosphoinositide 3-kinases.
NOTES
*Volutin in Saccharomyces cerevisiae.