1. INTRODUCTION
The genus “Barbus” is a complex one with members distributed widely, occuring in almost every freshwater. The genus comprises of more than 800 species worldwide and about 50 of them are reported from Africa [1]. In Tanzania, the genus barbus is represented by 37 species including: Barbus jacksonii, B. paludinosus and B. trimaculatus by name, but few are found in different habitats such as rivers, streams and Lakes. The genus has high morphological plasticity [2], and contains different levels of ploidy [3] which in most cases delimits identification of the members of the genus “Barbus”. For instance, [4] clearly splits the “Barbus species” into five mitochondrial lineages corresponding to Barbus sensu stricto (tetraploid, which is further subdivided into subgenus Barbus and Luciobarbus), the hexaploid species, the Ethiopian tetraploid species, the African diploid species and the Asian diploid species. On the other hand [5] points out that the genus Barbus includes only the species from Europe, North Africa and Southwestern Asia (peri-Mediterrenean Barbus), and thus the Tropical and South African species (other than those from North Africa) belong to several other genera yet to be taxonomically named. Henceforth, [5] and Skelton (per. Com., 2012) concluded that the current systematic of the genus Barbus can no longer be maintained. However, more barbus species needs to be studied prior to introducing new nomenclature.
This study therefore, aimed at estimating the phylogenetic relationship among the “Barbus species” occurring in Malaragasi and Pangani river basins years posed uplifting of the East African topology, and related the phylogeny to the wider context of the genus elsewhere in Africa, south of the Sahara. This was achieved through the partial sequencing of mitochondrial cytochrome b gene. The cytochrome b gene has, in the past, provided complementary and informative sequence data sets for determining phylogenetic relationships between morphologically similar species. Therefore, it was expected to be a suitable phylogenetic marker to address the questions posed in this study.
2. MATERIALS AND METHODS
2.1. Study Area
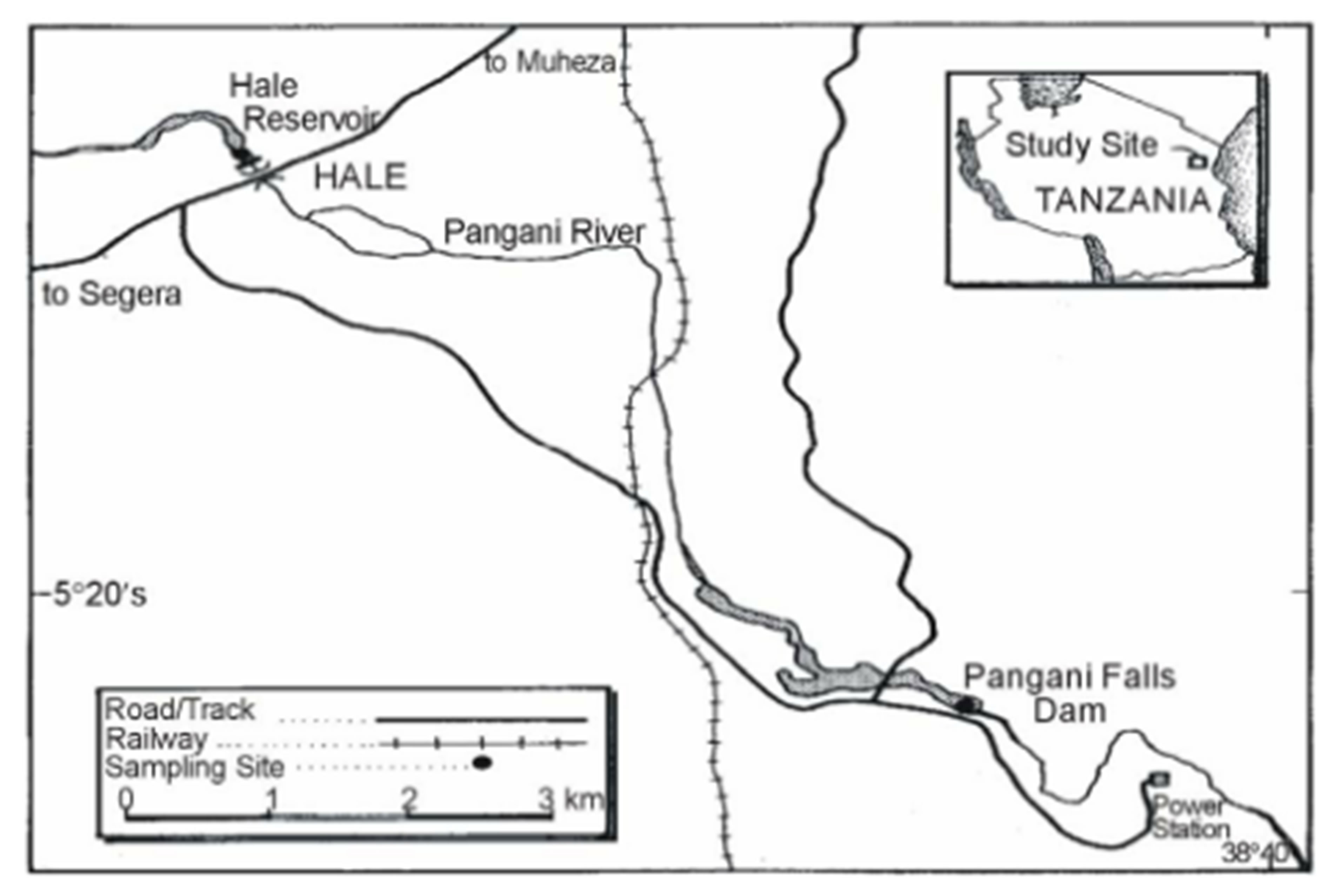
The study was based in Pangani and Malagarasi river basins. The Pangani basin is located in the North Eastern side of Tanzania between latitude 3˚03'S and 5˚59'S and longitude 36˚23'E and 39˚13'E with an area of about 53,600 km2 (Figure 1(a)). The basin is composed of five river sub-catchments, namely Pangani, Zigi, Umba, Mkulumuzi and Msangazi with main rivers draining to the Indian Ocean. Lakes in the basin are Chala, Jipe, Duluti, Manga and Karamba. Nyumba ya Mungu, Mabayani and Kalimawe Dams are manmade reservoirs. On the other hand, the Malagarasi basin is located in the North Western side of Tanzania (Figure 1(b)). Samples were collected from Lakes Sagara and Nyamagoma within the Malagarasi-Muyovozi Wetland systems. The lakes are located 200 km North East of Lake Tanganyika. The sites were chosen due to the fact that they are located in the Malagarasi-Muyovozi Ramsar site which is the first Ramsar site in Africa since 2000. Secondly, the Malagarasi River is one of the Potential fish biodiversity hot spots in Africa [6].
2.2 Collection of Fish Samples
In total 46 individuals of Barbus species from Pangani and Malagarasi rivers, plus 24 DNA sequences of the same species from Mozambique, Zambia, Congo and Malawi used in previous studies obtained from SAIAB’s collection were examined. The fishes were caught by traps, gillnets, monofilaments and some of them were obtained from local fishers.
2.3. Sample Preservation
The obtained samples were soaked into 99.8% alcohol contained in zip bag. The zip bags were placed into the plastic bags and stored in the buckets for easy carrying and transport. In the laboratory, the samples were transferred into zip rock containing 60% - 70% alcohol and stored at room temperature.
2.4. DNA Sources and Extraction
Genomic DNA was extracted from the muscles (on one left side) of each specimen following the protocol Wizard Genomic DNA Purification Kit (Promega). In addition, some other analysis materials were obtained from the collection at South African Institute of Aquatic Biodiversity (SAIAB).
 (a)
(a) (b)
(b)
Figure 1. Map of Tanzania showing the sampling points at A: Malagarasi and B; Pangani river basins.
2.5. PCR Amplification, Cloning and Sequencing
Two primers (Glu-F, 5’-GAA GAA CCA CCG TTG TTA TTC AA-3’) and (M93R,5’-GAA GAA CCA CCG TTG TTA TCA A-3’) were used to amplify cyt-b gene via polymerase chain reaction (PCR) [7]. PCR reactions were carried out in 50 µl reaction containing 5 µl dNTP (1 mM), 5 µl (10×) reaction buffer, 5 µl (25 mM) MgCl2, 2 µl (10 mM) each primer, 5 µl total DNA, and 0.3 µl Taq DNA polymerase. PCR cycles for amplications were performed using the following conditions: 35 cycles of denaturation at 94˚C for 2 minutes, annealing at 56˚C for 45 minutes, and extension at 72˚C for 2 minutes and 30 seconds. A final extension at 72˚C for 5 minutes was performed to completely extend the amplified product and the reactions were held at 4˚C for an average of 5 minutes. The PCR products were purified with the QIAquick (QIAGEN) kit following manufacturer’s protocol. Each PCR product was sequenced using the BigDye Terminator cycle sequencing ready reaction kit (Applied Biosystems Inc.) on an automated DNA sequencer (Applied Biosystems 377), following manufacturer’s instructions.
2.6. Phylogenetic Analysis
DNA sequences were aligned based on the inferred cytochrome b amino acid sequences. No ambiguous alignments were found and no gaps. All codon positions were included in the phylogenetic analyses. There were a total of 870 positions in the final dataset, out of which 439 were parsimonious informative. Phylogenetic analyses were conducted in MEGA4 [8]. The cytochrome b data set was subjected to the maximum parsimony (MP) method (PAUP* version 4.0 b2a [9] using heuristic searches 1000 replications (TBR branch swapping; MULPARS option in effect) with hundred random stepwise additions of taxa to find the most parsimonious trees. The robustness of the inferred trees was tested by bootstrapping [10,11] using 10,000 replications.
3. RESULTS
Three trees were derived with MP method separating the barbs of Malagarasi and Pangani basins into lineages corresponding to their respective localities. However, there were some striking findings, one individual Barbus paludinosus from the Hale separated from the rest of the group. Similarly the B. paludinosus from Hale completely separated from those of the Pangani falls though sharing the same catchment. The same findings were noted for the Muumbara and Songati populations. Two lineages were also observed for the B. trimaculatus population of Lake Nyamagoma, with one of the group coming closer to B. jacksonii of the Pangani falls (Figure 2).
When sequences of Barbus specimen (SAIAB collection) from southern part of Africa were included in the analysis, the MP method yielded a tree that separated the Barbus paludinosus of Malagarasi and Pangani basins from those of Malawi and Zambia. Similarly Barbus trimaculatus and Barbus jacksonii from the two basins of Tanzania came out independently from those of Mozambique (Figure 3).
Figure 4 combined published sequences of different Barbus species from various African countries and Central Europe obtained from the geneBank for comparison. Cyt.b sequences separated Barbus species into the clade of African Barbus species and the European Barbus species. In this relationship, Barbus paludinosus is highly related to Barbus pleurogramma from L. Tana Ethiopia (93% bootstrap values). The South African redfins such as P. asper, P. quanthlambae, P. burchelii and P. burgi believed to be closely related to small African Barbus species clustered together to form their own clade (78% bootstrap values). The Europe species of the genus Barbus and other related species formed their own clade