Optimal Estimation of High-Dimensional Covariance Matrices with Missing and Noisy Data ()
1. Introduction
The covariance matrix is a key component in various fields, particularly statistics. However, when dealing with many statistical situations, the covariance matrix is usually unknown. As a result, estimating the covariance matrix is extremely significant, and it is frequently utilized in signal processing, genomics, financial mathematics, and other domains. When the dimension p is fixed and the sample size n is sufficiently large, the sample covariance matrix is commonly used to estimate the true covariance matrix. However, with advancements in information technology and various other technologies, there is a growing challenge in estimating large covariance matrices. Issues such as dimensionality and noise can significantly impact the effectiveness of using the sample covariance matrix to estimate the true covariance matrix. Moreover, in the era of big data, missing data is a common occurrence, making research on the estimation of high-dimensional covariance matrices based on missing and noisy data essential.
Bickel and Levina [1] proposed thresholding as a commonly used method for estimating high-dimensional covariance matrices and and proved its consistency under the operator norm. However, there was no discussion on its optimality. Cai and Zhou studied the optimal estimation of sparse covariance matrices under the operator norm and Bregman divergence loss. They also proved that the thresholding estimator can achieve the optimal convergence rate under the spectral norm, see [2] . Cai and Zhou [3] provided the optimal estimation of the sparse covariance matrices under the
norm loss. The thresholding described above is also referred to as hard thresholding, and its counterpart is soft thresholding [4] [5] . On this basis, Rothman, Levina, and Zhu [6] proposed generalized thresholding and proved its consistency. Cai and Liu [7] proposed adaptive thresholding. The adaptive estimation of high-dimensional sparse precision matrices was studied by Cai, Liu, and Zhou [8] . For bandable covariance matrices, [3] , [9] , and [10] conducted in-depth research.
In the case of missing data, Cai and Zhang [11] assumed that the missingness was independent of the data values and studied the optimal estimation of two classes of covariance matrices. Qi [12] explored the optimal estimation of sparse covariance matrices under the
norm and the Fribenius norm, respectively. In addition, the lower bound for estimating bandable covariance matrices under the spectral norm was studied based on noisy and missing data, but its optimality is not considered. Shi [13] studied the optimal estimation of bandable covariance matrices based on missing and noisy sample data.
It is not difficult to find that the research on estimating high-dimensional covariance matrices is primarily based on complete data. However, the correlational research on missing data and noisy models remains critical. The articles listed above served as a tremendous source of inspiration for this paper’s study topic and methods. This paper will provide corresponding research for the aforementioned issues. Sparse covariance matrices are widely employed in a variety of applications, including genomics. As a result, it is necessary to investigate the estimate of this kind of covariance matrix. The research in this paper can help people better estimate the high-dimensional covariance matrix when the sample is noisy and missing. Thus, it is convenient for many fields to better use high-dimensional data to obtain more useful information, and this paper provides them with a reliable theoretical basis.
The remaining sections of this paper are as follows: Section 2 will provide the associated concepts and knowledge of covariance matrix estimation, which serves as the theoretical foundation for the research. In Section 3, we will study the optimal estimation of sparse covariance matrices with missing and noisy data. In Section 4, numerical simulation experiment will be performed to investigate the estimating effect of the estimator presented in Section 3. The fifth section summarizes the research content and discusses existing problems.
2. Theoretical Basis
This paper will primarily study the optimal estimation of covariance matrices under the
norm. For a matrix
,
represents the singular values of
, while
represents the eigenvalues of
. The operator norm of
is defined as
There are three common operator norms:
(1)
norm:
;
(2) spectral norm:
;
(3)
norm:
.
Next, we will introduce the sub-Gaussian random vector. If there is a parameter
such that
, the random variable
is considered a sub-Gaussian random variable with parameter
, that is,
. It is easy to know that sub-Gaussian random variables include Gaussian random variables whose mean is 0 and all bounded random variables with a mean of 0. Assuming the random variable
is sub-Gaussian, its sub-Gaussian norm is denoted by
A p-dimensional random vector
is called the sub-Gaussian random vector if any linear combination of
is sub-Gaussian. That is, when
, for any
,
, and
, there is
Assume that a p-dimensional random vector
has the mean
and the covariance matrix
. Covariance matrix estimation is the process of computing a covariance matrix
based on
independent copies
of
and then using
to estimate
, i.e., making
approximate
in a certain sense. In this paper, minimax risk is used as a standard to measure the estimation effect. Suppose
has a certain class of covariance matrices, and
is a specific collection of
’s distributions. Then, under the specified matrix norm
, [3] defines the minimax risk of estimating
over
as
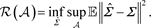
When the vector’s dimension
is smaller than the sample size
, the sample covariance matrix is typically utilized to estimate the true covariance matrix. The sample mean is
and the sample covariance matrix is
(1)
However, as noted in Section 1, when the dimension
is substantially larger than the sample size
, utilizing the sample covariance matrix for estimating the true covariance matrix becomes inadequate. Based on the work of Cai et al., this paper will study the optimal estimation of high-dimensional covariance matrices based on missing and noisy data.
The missing completely at random (MCR) model is introduced below.MCR indicates that the missingness was random and independent of the data values. Suppose
is complete random sample from
. Introducing vector
as the observation index for
, then
and
represent the
th coordinate of vectors
and
, respectively.
We denote
as the sample with missing data, where
is the
th observation sample. Additionally, define
When
, the
th and
th components of vector
are observed simultaneously, whereas
indicates that they were not observed simultaneously. Thus,
denotes the number of times the
th and
th components of
are simultaneously observed. For convenience, let’s define
,
. It is simple to know that
. When the sample data are complete,
.
For sample
with missing data, we substitute the generalized sample mean and generalized sample covariance matrix for the traditional sample mean and covariance matrix. The generalized sample mean is defined as the following:
the generalized sample covariance matrix is defined as
(2)
3. Covariance Matrix estimation
This paper assumes that the covariance matrix is sparse, which means that the majority of its components are 0 or insignificant, and the distribution of non-zero elements is irregular. First, we introduce the parameter space
of the sparse covariance matrices:
,
where
, and
represents the element with the
th largest absolute value in the
th column of matrix
. When
, each column of the matrices in
has at most
non-zero components, usually assuming
.
3.1. Noisy Model
Assuming the complete random vector
has the covariance matrix
. Using a
-dimensional random vector
to represent noisy data, the noisy model can be expressed as
(3)
where
represents noise. In this section, we hope to build a
matrix
based on
independent random noisy samples
of
. We next use
to estimate the covariance matrix
of the random vector
.
The noisy sample with missing data are represented by
, where
is the
th observation sample. The definition of the generalized sample mean is as follows:
the generalized sample covariance matrix is defined as
(4)
Two new assumptions in [12] are presented below.
Assumption 1. The observation index
can be random or deterministic, but it is independent of the noisy observation sample
.
Assumption 2. The random vectors
are i.i.d., where
, and
represents a fixed
-dimensional mean vector.
are fixed matrices with
and
. Each component of the random vector
i.i.d. sub-Gaussian with a variance of 1 and a mean of 0. For any
, there exists a parameter
such that
, that is,
.
3.2. Upper Bound for Estimating Sparse Covariance Matrix
The hard thresholding estimator based on complete data was proposed by [1] . When most of the elements in each row or column of the true covariance matrix are close to zero or negligible, set the elements of the sample covariance matrix below a certain threshold to 0, and leave the remaining elements unaltered to estimate the true covariance matrix, so as to reduce the error. In [2] ,
for
, where
is a constant. The threshold is set to
.
In this paper, it is extended to the case of missing and noisy data. According to Lemma 4.6 in [12] , if Assumption 1 and Assumption 2 are both hold, then there are two absolute constants
and
greater than 0, such that
(5)
for any
. Since
, the above can be simplified to: there are constants
and
, such that
(6)
Where
, and the constants
and
only depend on
. Note that Inequality can be written as
when
.
The hard thresholding estimator
of the covariance matrices
is defined by transforming the generalized sample covariance
in Equation (4),
(7)
where
is a constant and
.
The following is Lemma 1, which plays an important role in studying the minimax upper bound. Lemma 1 generalizes Lemma 8 in [8] from complete to noisy and missing sample.
Lemma 1. Define event
, then there is constant
, which only depends on
, such that
Proof : Firstly, define event
. It is easy to know that
(8)
According to the definition of
in Equation (7),
(9)
Next, we will prove this lemma in different cases. It can be obtained by simple calculation:
Therefore, there exists a constant
, such that
.
Next, we can obtain the upper bound for estimating the sparse covariance matrices by utilizing the risk properties of thresholding estimator.
Theorem 1. If Assumption 1 and Assumption 2 hold,
and
, then there is a constant
such that the hard thresholding estimator
defined by Equation (7) satisfies
(10)
Proof: Easy to know,
. If event
occurs,
Simple calculations show that
According to the definition of
, we know that
, so
. Select the constant
to satisfy
, so
Let the matrix
satisfy
, we have
Then, it is straightforward to acquire
Therefore, we only need to prove that
is negligible.
Firstly,
According to the Cauchy-Schwartz inequality, we know that
, and because
,
, so
In addition,
. From Inequality (5),
From Lemma 1, we have
and
, so
To sum up,
.
3.3. Lower Bound for Estimating Sparse Covariance Matrix
Before studying the lower bound, introduce some useful lemmas and symbols.
Lemma 2. Assume
and
are two probability measures, with
and
representing their probability density functions. The total variation distance between
and
is
. Define the total variation affinity as
. The Kullback divergence between
and
is expressed as
. Thus,
and
satisfy the following inequality:
(11)
Lemma 2 in [14] and Le Cam’s lemma and its corollary in [2] [12] introduced below are important tools for proving minimax lower bound.
Lemma 3 (Le Cam). Suppose
is a finite set of parameters. Let
be a loss function and
, then
is any estimator of
based on the observed values of the probability measure
, and
.
Lemma 4. Suppose
be any estimator of
based on the collection of probability measures
. We get
where
.
Before studying the minimax risk lower bound, it is advisable to construct a matrix with all off-diagonal elements equal to 0 except the first row or column. Let
be the collection of
symmetric matrices in which exactly
non-diagonal elements in the first row or column equal to 1 and all other elements are 0. Let
. Define
(12)
where
represents the identity matrix of size
,
, and
is a constant. Assuming
,
, it is easy to know that
.
Obtaining the lower bound requires two steps. Firstly, the subset of the parameter space constructed above is selected to simplify the proof. Secondly, calculate the total variation affinity between two probability measures.
Theorem 2. Let
,
, and
. Assume
with
. For any
, there exists a constant
such that the minimax risk lower bound for estimating the covariance matrix
satisfies
(13)
where
is any estimator of
based on noisy sample.
Proof: Assume
has
elements, where
represents the identity matrix and
represent the non-identity matrix, then
.
Assume that
, and the probability measure and probability density function are
and
, respectively, that is,
. Let
with
and
is the probability measure. Since
, it is easy to know that
Therefore, to prove Inequality (13), just prove the following Inequality:
(14)
Lemma 3.3 shows that
Since
and
, there exists a constant
such that
(15)
Obviously, to prove Inequality (14), we only need to prove that there is a constant
such that
.
From
, we have
Let
, it is easy to know that
. Suppose the eigenvalues of
are
, then there are
(16)
In addition, we can know that
(17)
where
is a number between 0 and
. Putting Equation (16) and Equation (17) into
, we can get
According to Theorem 1.3 in [12] ,
It is easy to see that
, hence
Lemma 2 implies
That is, there exists a constant
such that
.
It is worth noting that Theorem 2 requires
, which is a necessary condition. If
, then
does not have a consistent estimator in this case.
Theorem 1 and Theorem 2 show that the estimator
we construct is rate-optimal over
under the
norm.
4. Numerical Analysis
The optimal estimation of sparse covariance matrices based on missing and noisy data is derived in Section 3. This section compares the performance of the hard threshold estimator
, as defined in Section 3, against the traditional estimator using numerical simulation.
Some symbols are presented before the numerical simulation begins. Assume the
-dimensional Gaussian random vector
has a mean of
and a covariance matrix of
.
is the number of samples, and
is their dimension.
Here are the specific steps of numerical simulation.
1) Construct the sparse covariance matrix.
Assume
is a zero vector and
is a sparse matrix (
). Consult the construction of the sparse matrix in [11] , let
where
, and
,
,
.
2) Generate random samples according to the true covariance matrix.
After
is constructed,
-dimensional random samples are first generated from the multivariate normal distribution with mean
and covariance matrix
. The resulting
samples are then subjected to noise with a sub-Gaussian distribution, followed by random missing processing. This method produces sample data with missing and sub-Gaussian noise.
3) Compare the estimation effect of different estimators.
Based on the sample data with missing and sub-Gaussian noise, calculate the generalized sample covariance matrix
and the hard thresholding estimator
according to Equation and Equation . Then compute the error between
and the real matrix
, as well as the error between
and the real matrix
, under the given norm.
After determining the values of
and
, repeat the above three steps 1), 2), and 3) 50 times, and take the mean value of the fifty error results as the standard for evaluating the estimation effect of different estimators in this case. The performance is better when the outcome is smaller. Table 1 shows the experimental results.
The values of
and
are shown in the first two columns of Table 1. Table 1 shows the average after 50 runs of the processes 1), 2), and 3) with
and
fixed. When the true covariance matrix
is sparse, the hard thresholding estimator
has a substantially better performance than the generalized sample covariance matrix
under any norm, especially when
is larger than
, that is, the dimension is high.
Therefore, when the dimension is very small in comparison to the sample size, the sample covariance matrix can be used to estimate the population covariance matrix. When estimating a high-dimensional sparse covariance matrix with sub-Gaussian additive noise and missing data, it is best to choose the hard thresholding estimator
given in Equation (7). This section provides some suggestions for application statisticians on how to select estimation methods.
![]()
Table 1. Results of estimating sparse covariance matrix.
5. Summary and Outlook
In statistics and other fields, covariance matrix estimation is crucial. The estimation of high-dimensional covariance matrices has always been a hot topic with the rapid growth of numerous technologies.
Based on the missing and noisy sample data, this paper constructs a hard thresholding estimator
, and studies its optimality. Section 3 shows that the hard thresholding estimator given in this paper is rate-optimal. The numerical simulation shown in Section 4 demonstrates that the hard thresholding estimator works well in situations where the true covariance matrix is sparse. When the true covariance matrix is not sparse, the estimation effect of the hard thresholding estimator has not been discussed.
This paper’s research has limitations and areas that require more investigation:
1) This paper focuses solely on the optimal estimation of sparse covariance matrices based on noisy and missing data. More research is needed on the optimal estimation of other common high-dimensional covariance matrices.
2) If the sub-Gaussian distribution used in this article is replaced with the sub-exponential distribution with a larger range, the relevant issues merit additional investigation.