1. Introduction
Diarrhoea is the second leading cause of death among children < 5 years of age. It accounts for one out of twenty seven child fatalities globally, with 80% of these occurring in low-middle-income countries [1] . In Kenya, diarrhoea is responsible for 17% of all childhood diseases, with children < 5 years experiencing, on average, three incidences of diarrhoea annually [2] [3] . The common etiologies of diarrhoea are bacteria, viruses, and parasites. Among bacteria, diarrheagenic Escherichia coli (DEC) strains are responsible for 30% - 40% of acute diarrhoea episodes, and drive periodic infections and diarrheal outbreaks worldwide [2] . The introduction of the rotavirus and cholera vaccines reduced the burden of diarrhea in children by 52% - 59% globally [4] [5] . However, bacteria-associated diarrhea remains a problem in informal settlements, which have poor sanitation and hygiene, and variable access to vaccines.
Five common DEC pathotypes with distinct virulence and pathogenic characteristics have been described. These include enteroaggregative E. coli (EAEC), enterohemorrhagic E. coli (EHEC), enteroinvasive E. coli (EIEC), enteropathogenic E. coli (EPEC), and enterotoxigenic E. coli (ETEC) [6] . These are associated with, among others, travellers’ diarrhoea, haemorrhagic colitis, haemolytic-uremic syndrome and acute and persistent diarrhoea [6] .
Antibiotic-resistant DEC isolates have been reported globally complicating the management of bacterial diarrhoea [7] . In Kenya, the first multidrug-resistant EAEC infection was reported in Malindi in 1997 [8] and is currently widely disseminated. Recent studies have reported high rates of resistance to ampicillin (AMP) and sulfamethoxazole/trimethoprim (SXT) among DEC strains [9] , indicating increased transmission and the possible emergence of novel resistant strains. Nevertheless, frequent monitoring of DEC pathotypes is not conducted, and diarrhoea infections are treated empirically, potentially enhancing the selection of resistant strains. Identifying these pathogens is critical, particularly in informal settlements where diarrhoea outbreaks are commonly reported [10] , and where inadequate sanitation, poor hygiene and limited access to clean water can enhance their transmission and maintenance [11] . This study investigated the DEC pathotypes associated with acute diarrhoea infections and their antimicrobial susceptibility profiles among children attending three outpatient healthcare facilities within the Mukuru informal settlement in Nairobi, Kenya, to guide the management of E. coli-related diarrhoea.
2. Materials and Methods
Study population. We analysed archived E. coli isolates from stool samples of children aged < 5 years, who presented with diarrhoea at three outpatient clinics—Mukuru Kwa Njenga, Mukuru Kwa Reuben, and Municipal County Council—located within the Mukuru informal settlement in Nairobi County. These children were enrolled in an enteric fever surveillance study [12] . E. coli isolates collected between January 2017 and September 2018 were included.
Isolate revival. Isolates were retrieved from −80˚C freezer and thawed at room temperature for 20 min. These were then plated on MacConkey agar (Oxoid, UK) and incubated at 37˚C for 18 h. Single colonies were obtained from each plate and sub-cultured on Mueller Hinton agar (Oxoid, UK) at 37˚C for 18 h. Confirmatory identification of pure isolates was done using the VITEK®2 Gram-negative identification cards (Biomerieux, France).
DNA extraction. Once confirmed as E. coli, isolates were sub-cultured overnight on Mueller Hinton agar. Single colonies were collected from each plate and emulsified in 1 mL of Invitrogen DNase/RNase free water (Thermo Fisher Scientific, USA) in a sterile microcentrifuge tube and boiled at 95˚C for 12 min. Upon cooling, the suspension was centrifuged for 5 min at 15,366 ×g and 200 µL of the supernatant containing DNA transferred into a sterile Eppendorf tube for downstream analyses.
Molecular analysis. The Applied Biosystems PCR machine (Thermo Fisher, USA) was used to amplify genes coding for elt (heat labile enterotoxins), est (heat stable enterotoxins), stx (shiga toxin), aggR (activator aggregative adherence regulator), aspU (secreted protein U gene), ipaH (invasion plasmid antigen), and eae (intimin) using select primers (Table 1). Polymerase chain reaction (PCR) was done following the protocol by Toma et al. [13] , i.e., initial denaturation at 94˚C, 30 s, 30 cycles of amplification at 94˚C for 30 s, annealing at 55˚C (EPEC), 53˚C (EAEC), 58˚C (EHEC), 56˚C (ETEC), 66˚C (EIEC) for 1 min, initial extension at 72˚C for 2 min, and final extension at 72˚C for 7 min. Clinical isolates containing DEC virulence genes were used as positive controls and E. coli ATCC 25922 was used as a negative control [10] .
Amplified PCR products were run on a 2.5% Hi-Res Standard Agarose gel (Agarose 0.5 × Tris-borate-EDTA) for 1 h. The gel was stained using 0.5 µg/mL ethidium bromide and visualized under an ultraviolet illuminator. PCR amplicon sizes were confirmed using a 1 kb Invitrogen ladder (Thermo Fisher, USA).
Antibiotic susceptibility testing. All DEC isolates were tested for susceptibility to 17 antibiotics—belonging to 11 antibiotic classes [16] —using the VITEK®2 AST-GN71 cards. Interpretation of antibiotic susceptibility results was based on the 2020 Clinical and Laboratory Standards Institute (CLSI) guidelines [17] . Quality control testing for the VITEK®2 cards was done using manufacturer
![]()
Table 1. Diarrheagenic E. coli pathotypes PCR primer sequences.
Enteroaggregative-(EAEC), enterohemorrhagic-(EHEC), enteroinvasive-(EIEC), enteropathogenic-(EPEC) and enterotoxigenic E. coli (ETEC). ᶲ [13] , † [14] , * [15] .
recommended ATCC strains. Multidrug resistance was defined as resistance to at least one antibiotic in three or more antibiotic classes.
Data analysis. Data was analysed using frequency distributions and proportions for pathotypes and AST results on SPSS version 21 and Microsoft Excel Spreadsheet Software (2016).
Ethics statement. This study was approved by the Kenyatta National Hospital-University of Nairobi Ethics and Research Committee (P512/09/2020).
3. Results
Diarrheagenic E. coli pathotypes
Of the 175 E. coli isolates analysed, 48 (27%) were pathogenic including 34 EAEC (71%), nine EHEC (19%), and five EPEC (10%). EIEC and ETEC pathotypes were not identified. Among EAEC isolates, only the aggR gene was detected.
Antibiotic susceptibility profiles of E. coli pathotypes
All DEC isolates were fully susceptible to amikacin, ertapenem, imipenem, meropenem and tigecycline. Most (>80%) were resistant to ampicillin, ampicillin-sulbactam and sulfamethoxazole-trimethoprim. All EAEC and EPEC isolates were resistant to cefazolin while EHEC where < 50% of isolates were. In general, <30% of EAEC and EHEC isolates were resistant to ceftriaxone, cefepime, gentamycin and tobramycin; EPEC strains were susceptible to these antibiotics, and ciprofloxacin. All EHEC isolates were susceptible to nitrofurantoin (Figure 1).
Resistance phenotypes
In total, 18 resistance phenotypes were identified among the 48 isolates (Table 2). The most common resistance phenotype was “AMP-CEZ-SAM-SXT”, which
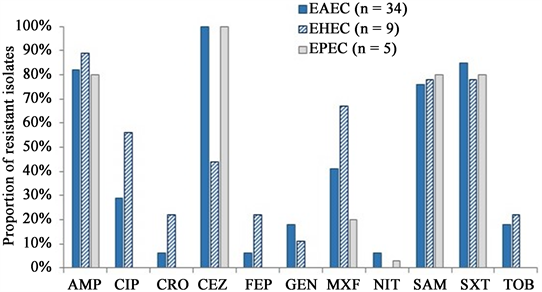
AMP, ampicillin; ATM, aztreonam; CIP, ciprofloxacin; CRO, ceftriaxone; CEZ, cefazolin; FEP, cefepime; NIT, nitrofurantoin; GEN, gentamicin; MXF, moxifloxacin; SAM, ampicillin-sulbactam; SXT, sulfamethoxazole-trimethoprim; TOB, tobramycin.
Figure 1. Proportion of resistant diarrheagenic E. coli pathotypes identified among children < 5 years old in the Mukuru informal settlement, Nairobi.
![]()
Table 2. Resistance phenotypes of diarrheagenic E. coli isolates.
Enteroaggregative-(EAEC), enterohemorrhagic-(EHEC), enteroinvasive-(EIEC), enteropathogenic-(EPEC) and enterotoxigenic E. coli (ETEC). AMP, ampicillin; ATM, aztreonam; CIP, ciprofloxacin; CRO, ceftriaxone; CEZ, cefazolin; FEP, cefepime; NIT, nitrofurantoin; GEN, gentamicin; MXF, moxifloxacin; SAM, ampicillin-sulbactam; SXT, sulfamethoxazole-trimethoprim; TOB, tobramycin.
was observed in 33% of isolates, and was the only phenotype identified in all three pathotypes. “AMP-CIP-CEZ-GEN-MXF-SAM-SXT-TOB” and “CEZ” were the next most common phenotypes each accounting for a 10% representation. Overall, 81% of all DEC were multidrug-resistant. Four (8%) of the 48 isolates tested positive for ESBL-production two EAEC (6%, n = 34) and two EHEC (22%, n = 9) isolates.
4. Discussion
Diarrheagenic E. coli are among the primary causes of diarrhea in low-middle-income countries [15] [18] . The overall diarrhea burden associated with DEC is not well defined in Sub-Saharan Africa due to poor testing infrastructure [19] . This study reports a proportion of 27% of DEC, higher than what was previously reported in four provinces in Kenya, Zambia and South Africa [11] [20] [21] . However, this is lower than the proportions reported in Uganda and central regions of Kenya [22] [23] . The distribution of DEC pathotypes varies geographically due to seasonal variations, animal interactions and poor sanitation and hygiene [9] [11] [24] . Moreover, host factors such as malnutrition and HIV status are associated with some DEC pathotypes [22] contributing to the regional and global variation of these pathotypes. The most effective therapy for DEC-related diarrhea is vaccination, but the immunological heterogeneity of the strains has, for instance, challenged the development of ETEC vaccine [25] .
Three E. coli pathotypes were associated with diarrhoea among children aged < 5 years in our study population. EAEC was the common pathotype, consistent with previous studies in Kenya and Uganda [9] [22] [24] . EAEC is an emerging pathogen responsible for acute and persistent diarrhea across all ages worldwide [2] [26] [27] , although a higher prevalence is reported among young children in developing nations [28] . Poor sanitation, inadequate water supply and compromised hygiene—conditions that are prevalent in informal settlements and which promote recurrent diarrheal episodes among children—are some of the risk factors associated with EAEC infections in children [9] [29] . Further, EAEC strains may persist in water for almost 60 days at standard storage temperatures, enhancing their transmission [29] .
EHEC, which was the second most common pathotype, is typically a zoonotic food-borne pathogen which causes bloody diarrhea and kidney failure through the expression of a Type III secretion system and Shiga toxins, respectively [30] . In India, living in close quarters with domestic animals has been associated with increased risk of EHEC infections in humans [31] . Environmental transmission could also be a source for EHEC infection in children especially during their explorative stages [32] . In the Mukuru informal settlement, only 23% and 12% of households keep cats and dogs, respectively, with <10% owning livestock, suggesting that contact with domestic animals may not be a significant risk factor for EHEC infections in this setting [33] . Nevertheless, its comparatively lower detection relative to EAEC may indicate low-level exposure to animal faeces within the environment or an alternative exposure.
EPEC was the least reported pathotype in this study contrary to earlier studies done in Kenya, Nigeria, India, and Qatar [20] [34] [35] . EPEC is a recurrent agent of diarrhoea and is associated with high mortality among children age < 11 months [36] [37] . A previous study in four counties in Kenya reported an overall prevalence of 51% among children < 5 years [20] suggesting that its epidemiology might be changing. This may explain the decreasing number of reports of its occurrence in industrialized countries than initially described [38] . In Kenya, the changing epidemiology may be attributed to overall improvements in water, sanitation, and hygiene, advocacy for exclusive breasting among infants and the introduction of zinc supplements for the management of childhood diarrhea [9] .
More than half of all DEC pathotypes were resistant to ampicillin, ampicillin-sulbactam, cefazolin and sulfamethoxazole-trimethoprim, consistent with previous studies in Kenya and Nigeria [2] [9] [39] . Most of these antibiotics are inexpensive oral formulations that are easily accessible and may thus be frequently used within this population. Further, most EHEC isolates were resistant to ciprofloxacin, an antibiotic to which increasing resistance has been reported among Salmonella sp. isolated in the same study area. This suggests possible gene transfer between bacteria in this setting and, possibly, antibiotic selective pressure [40] . In Kenya, antibiotic therapy for infant or childhood diarrhoea is recommended for the treatment of dysentery and salmonellosis, while oral rehydration solutions and zinc sulphate are recommended for the management of DEC-related diarrhoea [41] . However, low diagnostic capacities in low-middle income countries promote empiric treatment of childhood diarrhoea with antibiotics, which likely contributes to the emergence of resistant strains [42] . For informal settlements, where dense populations and unsanitary environmental conditions can facilitate the transmission of multidrug-resistant bacteria [43] [44] [45] , antibiotic stewardship efforts are best complemented with water, sanitation and hygiene interventions to reduce childhood diarrhea-related deaths.
We observed resistance—albeit low—to aminoglycosides (gentamicin and tobramycin), fluoroquinolones (ciprofloxacin and moxifloxacin) and a 4th-generation cephalosporin (cefepime) among EAEC and EHEC isolates, in contrast to studies in Kenya and Burkina Faso that reported complete susceptibility to these antibiotics [9] [46] . Further, we found a low occurrence of ESBL-positive DEC pathotypes (in EHEC and EAEC only) unlike in the Burkina Faso and Uganda studies [22] [46] . Even at low proportions, ESBL-producing E. coli are an important cause of life-threatening infections in informal settlements, whose spread is facilitated by poor sanitation and hygiene, hospitalization and indiscriminate antibiotic use which are prevalent is these settings [47] . Their occurrence, therefore, present a public health threat that necessitates interventions to curb further emergence and spread within communities.
The observed susceptibility to carbapenems and amikacin may be due to limited access to these antibiotics as they are not prescribed at outpatient clinics in Kenya. However, misuse of locally available β-lactam antibiotics such as ceftriaxone, ampicillin and amoxycillin may select for carbapenemase-expressing strains. For instance, hospital and community colonization with carbapenem resistant Enterobacterales have been recently reported in Kenya [16] , suggesting changing antimicrobial resistance trends that requires active surveillance, monitoring and antimicrobial stewardship in the community and hospital set ups.
Multidrug resistance was common among the DEC isolates, as has been shown in studies in Kenya, Qatar, Iran and Burkina Faso [34] [48] [49] [50] . The observed resistance to penicillin, sulfonamide, cephalosporin, and quinolone drug combinations has been reported in other countries at variable levels [21] [50] . Interestingly, recent studies in Kenya that involve DEC isolates have not reported multidrug resistance phenotypes that include cephalosporin and quinolone drug combinations [39] [48] , suggesting the possible emergence of novel strains. In contrast, other studies in Africa have reported multidrug-resistant phenotypes that involved penicillin, sulfonamide, colistin and tetracycline drug combinations [9] [39] [46] . Since our antibiotic panel did not include colistin and tetracycline we did not identify these phenotypes and cannot, therefore, confirm their presence in our study population.
Our study had a few limitations. By using archived isolates, we were unable to collect other data (e.g., participant demographics, persistent vs. acute diarrhea, location, antibiotic use, or coinfections) that may have helped to explain the observed findings. Further, the isolates analyzed in this study were collected from sick children who presented themselves at the study facilities, who represent a non-random subset of the population living in the Mukuru informal settlement. Consequently, our results may not be generalizable the entire child population in this setting. Lastly, given that the DEC strains isolated may have been a secondary cause of diarrhea among the enrolled children, we were unable to conclusively assess the severity of these pathogens in this population. Longitudinal cohort studies that include a randomized population of children may be best suited to address these limitations.
5. Conclusion
This study found that EAEC contributes most to DEC-related diarrheal infections among children living in the Mukuru informal settlement in Kenya. The presence of multidrug resistant strains, particularly phenotypes that are resistant to cephalosporin and quinolone drug combinations, suggests that enhanced water, sanitation, and hygiene programs remain the best strategy for preventing the emergence and spread of diarrhoeal diseases in these settings. Understanding the epidemiology of diarrheal disease and improving antimicrobial resistance surveillance can help identify the etiological agents of diarrhea and identify newly emerging resistant strains before they disseminate widely. This will inform targeted treatment guidelines for better management of childhood diarrhea and form a basis for future vaccine interventions.
Acknowledgements
We acknowledge the Centre for Microbiology Research in KEMRI for providing the samples analyzed in this study, Washington State University Global Health-Kenya and the University of Nairobi for providing the resources to analyse these samples.