Antibiogram, Genomic and Phylogeny of Stool and Seafood Isolates from Some Cholera-Prone Coastal Communities in Rivers State, Nigeria ()
1. Background
Cholera disease remains a significant public health threat across the globe, primarily originating in coastal regions or communities that depend on streams, rivers, or other sources of untreated water supply [1] . Coastal waters in the Niger Delta region serve as a receptacle of all manner of wastes, both toxic and non-toxic. These wastes may include chemical, biological, fecal, and, physical wastes. Pollution of coastal water bodies is a very critical challenge worldwide, especially in developing countries. Studies have shown that pollution of water bodies is the leading cause of diseases and deaths all over the world [2] . Coastal communities in Rivers State as well as numerous waterways and estuaries have become the channel for the removal of human effluent and the ocean as a disposal ground, bringing about eutrophication [3] . Estuaries specifically get many contaminating chemicals through waterway sources and deposits from the environment, which eventually get to the coastal areas. A portion of the more harmful substances directly impact the strength of marine/coastal species and through this, also affect the food chain and the working of whole biological systems. Simultaneously expanded misuse of coastal and marine resources, including commercial fishing alters populations, diminishes biodiversity, and exposes communities to infectious diseases including cholera [3] . Cholera is a life-threatening water-borne disease caused by the toxigenic strains of Vibrio cholerae. The disease is characterized by a watery and intense form of diarrhea, bulky stool, acidosis, and hypo-volemic shock [4] . It is both endemic and epidemic in different parts of the world, especially in developing countries where sanitary management is very poor [5] . Pathogenicity of Vibrio spp depends on the expression of some virulence factors such as choleragen (CT) which is mobilized by the toxin-Coregulated pilus (TCP). These genes are encoded by the ctx and tcp genes respectively [6] . There is also the outer-membrane protein (Omp) gene. This is a virulence factor involved in stimulating the immune response through the induction of protective immunity and also contributes to bacterial pathogenesis through the facilitation of adaptability of the pathogenic strains. This virulence factor is encoded by the OmpW gene [7] . Although CT production encoded by the ctxAB genes determines the progression of diarrhea, the ability of Vibrio cholerae to cause disease depends on the combined functions of different other genes, including genes for colonization factors [4] . The virulence genes are demonstrated in clusters, though there are about two regions of V. cholerae chromosome where virulence genes are clustered, viz the CTX element and the TCP accessory colonization factor gene cluster also known as the TCP pathogenicity island [7] .
Identification of Vibrio cholerae has been by conventional isolation on selective agar medium accompanied by biochemical and serological tests [8] . However, nucleotide-based identification technique such as polymerase chain reaction (PCR) is important for proper identification as this technique amplifies and defines target sequences that are present within a source of DNA [5] . The technique is also designed to allow the selective application of a specifically targeted DNA sequence within a heterogenesis collection of DNA sequences [9] . Conventional identification and typing techniques that have been used for years include serotyping, biotyping, susceptibility testing, and phagetyping. These have been challenged because of their relatively poor sensitivity, and specificity [10] .
2. Methods
2.1. Study Design
The research design used in this study is descriptive. The study was done seasonally from February to November 2020. This period covers the wet and dry seasons. The reason is that cholera is usually influenced by season; hence seasonal determination of Vibrio species is very crucial. The sample collected included water from six different water bodies, sea foods from associated communities, and stool samples.
The water bodies were situated in coastal communities, two water bodies from each Senatorial District in Rivers State. The water bodies include Marine Base Jetty, Chuku-Ama River, Kaa waterside, Andoni River, Egbolom River, and Bonny River. The seafood collected was based on common and available seafood at the time of collection. Both fin food and shell foods were collected. General Hospitals and Health Centers situated in the coastal communities were visited with a well-structured questionnaire which was administered to each participant with the help of attending Health personnel. The participants that met the criteria were recruited in the study and their stool samples were collected and subjected to standard microbiological and molecular analysis. An antibiogram of isolated organisms was done and resistant genes were determined from the isolates obtained from stool and seafood samples. The isolates were also examined for virulence genes using molecular techniques.
2.2. Inclusion and Exclusion Criteria
All participants within the range of 1 - 65 years, who signed written consent, experienced diarrhea for the past 3 months, were not on any antibiotic drug, and inhabitants of the Coastal Communities under study were included in the study.
Individuals who met the inclusion criteria but did not give their consent were excluded from the study. Apparently healthy persons that signed the informed consent but did not show any sign of diarrhea were excluded from the study.
2.3. Sample Collection
Seafood: Different kinds of seafood were collected from the six coastal communities (Okrika, Ogu-Bolo, Andoni, Khana, Abua-Odual, and Bonny). The choice of seafood collected in each coastal community was based on the most common seafood consumed by the inhabitants in each study area. Seafoods were collected seasonally in the first period of sampling i.e. dry season (beginning of the year) total of fourteen seafoods were collected and including Periophthalmus papilio (mudskipper), anabara, oyster and periwinkle from Kaa waterside in Khana Local Government Area; Uca tangeri (feedler crab), Crassotrea gasar (oyster) and Tympanotonus fuscatus (periwinkle) from Andoni river; Cirripedia spp (barnacle) and oyster from Marine Base Jetty Okrika; Uca tangeri (feedler crab) and periwinkle from Chuku-Ama river in Ogu-Bolo; crab from Egbulom river in Abua and Uca tangeri and broken marriage fish from Bonny river. The collection of samples was repeated at the peak and end of the wet season in all the coastal communities.
The sea foods collected were arranged separately in sterile bags and arranged in cold chains and transported to the laboratory without delay for processing and analysis.
Stool Samples: A total of 400 watery and semi-formed stool samples were collected from subjects who met the required criteria. A clean dry wide mouthed screw cap universal container was properly labeled and given to each subject for the production of the stool sample. The samples were preserved in alkaline peptone water, packed in an ice pack, and transported to the laboratory for analysis.
2.4. Bacteriological Examination of Seafood
The sea foods, mortar, and pestle used for mashing were washed using sterile water, covered with foil, autoclaved, and dried in a hot air oven.
The shell foods like periwinkles, barnacles, oysters, and anabara were cut using a sterile saw. Each seafood was mashed separately, and one (1 g) gram of mashed seafood was added to 10 ml of sterile freshly prepared normal saline with a sterile spatula and labeled appropriately. Each tube was centrifuged at 3000 rpm for 5 minutes. After the centrifugation, 0.1 ml of the supernatant was dropped on Thiosulphate citrate bile salt agar (TCBS) and MacConkey agar plates with an automatic pipette and spread with spreaders respectively. Also, 0.1 g of deposit was placed on the TCBS plates and MacConkey agar and streaked using a glass spreader. Thiosulphate citrate bile salt agar was used because it is a selective medium for Vibrio species while MacConkey was used to detect other Enterobacteriaceae. The media used were prepared according to the manufacturer’s instructions. The plates were incubated at 37˚C for 24 hours. Normal saline was used for dilution so as to maintain the property of their natural environment. At the end of the incubation period, the growth obtained was subcultured on new plates by streaking and incubated again for 24 hours at 37˚C. This was done to obtain pure colonies. The pure colonies obtained were cultured on slant media (TCBS slant and nutrient slant) for further analysis.
2.5. Examination of Stool Analysis
One gram of each stool sample collected was inoculated into peptone water. The stool samples were further cultured from the peptone water into TCBS, Nutrient agar, and MacConkey agar. These plates were incubated for 24 hours at 37˚C.
At the end of incubation, the plates were examined for characteristic colonies. The isolates were also subjected to wet prep, gram staining, and biochemical and susceptibility testing. Suspicious colonies were subcultured in a slant and transported to the Molecular laboratory for genomic analysis.
2.6. Antibiotic Susceptibility Testing
The isolates were incubated in Luria Bertani medium at 37˚C for six hours. After this susceptibility testing was done using Muller-Hinton agar plates, following Kirby-Baur technique [11] .
The antibiotics used include 15 ug Azithromycin (Azm), 30 ug Cefatazime (ctx), 5 ug ciprofloxacin (CIP), 30 ug Amikacin (AK), and 10 ug Colistin (CT), 30 ug Ceftazidime (CAZ), 10 ug imipenem (IPM), 30 ug Gentamycin (CN) and 30 ug Amoxicillin-Clavulanic acid 2:1 (AMC). Each isolate had 2 plates of Muller-Hinton agar where five sets of antibiotics were carefully placed in them using sterile forceps. The plates were incubated for 24 hrs at 37˚C and the results were recorded according to the criteria of the Clinical Laboratory Standards Committee [12] and the multidrug-resistant strains were investigated for the presence of ESBL genes.
2.7. Amplification of 16SrRNA
The 16SrRNA genes of the bacteria isolated were amplified on ABI 9700 thermal cycler (Applied Biosystems, Inqaba South Africa). The forward and reverse primers used were 5'-AGAGTTTGATCTGGCTCAG-3' (27F, Barcode: C23F) and CGGTTACCTTGTTACGACTT-3' (1492R, Barcode: 52BD7) respectively. The amplification was done for 35 cycles at the final volume of 40 uL [13] . Other components used include X2 dream Taq Master Mix containing Taq Polymerase, DNTPs, Mgcl, and buffer; extract of DNA at a concentration of 50 ng and water. The conditions in which the PCR performed the amplification were as follows: Initial denaturation at 95˚C for 5 minutes, denaturation at 95˚C for 30 seconds, annealing at 52˚C for 30 seconds, extension at 72˚C for 30 seconds and final extension at 72˚C for 5 minutes. The amplicons were kept cool by the thermocycler at a temperature as low as 4˚C. At the end of the amplification process, the amplicons obtained were resolved at one percent agarose gel (1 g agarose in 100 ml 1× Trisboric acid buffer) at 130 volts for 25 minutes. The agarose gel electrophoretic outcome was visualized on a blue light transilluminator connected to a computer system for capture.
2.8. Amplification of Resistance Genes
Extended-spectrum Beta-lactam uses genes mediated by plasmid were amplified with their specific forward and reverse primers to detect the presence of resistant genes cefotaxime (CTX-M), sulfhydryl variable (SHV) gene, Temoniera (TEM) gene and Oxacillinase (OXA) gene.
2.9. CTX-M Gene
The primers CTX-MF 5'-CGCTTTGCGA TGTGCAG-3' (17 base units, Bar Code 54AOA) and CTX-MR 5'-ACCGCGATATCGTTGGT-3' (17 base units. Barcode 52AOB) at a concentration of 2.4M were used for thermal amplification of CTX-M gene from the isolates. A total of 35 cycles were run at a final volume of 40 microliters. The components added include X2 dream. Taq Master Mix, 50 mg extracted DNA as template and water. The conditions of polymerase chain reaction (PCR) successfully employed is as follows: initial denaturation at 96˚C for 5 minutes, denaturation at 95˚C for 30 seconds, annealing at 52˚C for 30 seconds, extension at 72˚C for 30 seconds, and final extension at 72˚C for 5 minutes. The amplicons were resolved on 1% agarose gel for 25 minutes at 120 v and visualized on a blue light transilluminator for a 500 base pair size [14] .
2.10. SHV Gene
SHV/F 5' CGCCTGTGTATTATCTCCCT-31 (20 bases, bar Code: 52AO6) and SHV-R 5'-CGAGTAGTCCACCAGATCCT (20 bases, Barcodes: S2A07) primers were used to amplify the SHV genes of plasmid on a thermal cycler ABI 9700 Applied Biosystem for 35 cycles at a final volume of 40 uL and primer concentration of 0.4 M. The PCR components were x1 Dream taq master mix, the extracted DNA template 950 mg, and DNase-free water. The conditions of PCR were the same as in CTX-M gene amplification. The amplicons were also resolved by 1% agarose gel electrophoresis and visualized on ultraviolet transilluminator for 25 minutes at 120 voltage for 200 base pairs size [14] .
2.11. TEM Genes
TEM genes expressed by the isolates were amplified on ABI 9700 Applied biosystem thermocycler for 35 cycles using TEM forward and reverse primers at final volume of 40 µl and primer concentration of 0.4 M. The primers include: TEM/F 5'-TTTCGTGCGCCCTTATTCC-3' (20 bases, Barcode: S2A14) and TEM/R 5'-ATCGTTGTCAGAAGTAAGTTGG-3' (22 bases, Barcode: S2A 15). The components and PCR conditions were same as that of CTX-M and SHV genes. The amplicons were resolved on 1% agarose gel at 120 v for 25 minutes and visualized on ultraviolate transilluminator for a 400 base pair and amplicon size.
2.12. OXA Genes
OXA genes from the isolates were amplified using the OXA-1F: 5'-AGCCGTTAAAATTAAGCCC-3' and OXA-1R: 5'CTTGATTGAAGGGTTGGGCG-3' primers on an ABI 9700 Applied Biosystems thermal cycler at a final volume of 40 microlitres for 35 cycles. The PCR mix included: the X2 Dream taq Master mix supplied by Inqaba, South Africa (taq polymerase, DNTPs, MgCl), the primers at a concentration of 0.4 M and 50 ng of the extracted DNA as template. The PCR conditions were as follows: Initial denaturation, 95˚C for 5 minutes; denaturation, 95˚C for 30 seconds; annealing, 47˚C for 30 seconds; extension, 72˚C for 40 seconds for 35 cycles and final extension, 72˚C for 5 minutes. The product was resolved on a 1% agarose gel at 120 V for 25 minutes and visualized on a UV transilluminator.
2.13. Amplification of Virulence Genes
The following virulence genes were amplified on an ABI 9700 Applied Biosystems thermal cycler.
2.14. Efflux System (AcrB) Gene
AcrB genes from the isolates were amplified using the AcrBF: 5'-ATCAGCGGCCGGATTGGTAAA-3' and AcrBR: 5'CGGGTTCGGGAAAATAGCGCG-3' primers on a ABI 9700 Applied Biosystems thermal cycler at a final volume of 40 microlitres for 35 cycles. The PCR mix included: the X2 Dream Taq Master Mix supplied by Inqaba, South Africa (Taq polymerase, DNTPs, MgCl2), the primers at a concentration of 0.4 M and 50 ng of the extracted DNA as template. The PCR conditions were as follows: Initial denaturation, 95˚C for 5 minutes; denaturation, 95˚C for 30 seconds; annealing, 60˚C for 30 seconds; extension, 72˚C for 30 seconds for 35 cycles and final extension, 72˚C for 5 minutes. The product was resolved on a 1% agarose gel at 120 V for 25 minutes and visualized on a UV transilluminator
2.15. Thermostable Direct Haemolysin (Tdh) Gene
Tdh genes from the isolates were amplified using the TdhF: 5'-CTGTCCCTTTTCCTGCCCCCG-3' and TdhR: 5'-AGCCAGACACCGCTGCCATTG-3' primers on a ABI 9700 Applied Biosystems thermal cycler at a final volume of 40 microlitres for 35 cycles. The PCR mix included: the X2 Dream Taq Master Mix supplied by Inqaba, South Africa (Taq polymerase, DNTPs, MgCl2), the primers at a concentration of 0.4 M and 50 ng of the extracted DNA as template. The PCR conditions were as follows: Initial denaturation, 95˚C for 5 minutes; denaturation, 95˚C for 30 seconds; annealing, 50˚C for 30 seconds; extension, 72˚C for 30 seconds for 35 cycles and final extension, 72˚C for 5 minutes. The product was resolved on a 1% agarose gel at 120 V for 25 minutes and visualized on a UV transilluminator
2.16. Sanger Sequencing of 16SrRNA
Sequencing was done using the BigDye Terminator kit on a 3510 ABI sequencer by Inqaba Biotechnological, Pretoria South Africa. The sequencing was done at a final volume of 10 ul, the components included 0.25 ul BigDye® terminator v1.1/v3.1, 2.25 ul of 5 x BigDye sequencing buffer, 10 uM Primer PCR primer, and 2 – 10 ng PCR template per 100 bp. The sequencing conditions were as follows 32 cycles of 96˚C for 10seconds, 55˚C for 5seconds and 60˚C for 4 minutes.
2.17. Phylogenetic Analysis
Obtained sequences were edited using the bioinformatics algorithm Trace edit, similar sequences were downloaded from the National Center for Biotechnology Information (NCBI) data base using BLASTN. These sequences were aligned using MAFFT. The evolutionary history was inferred using the Neighbor-Joining method in MEGA 6.0 [15] . The bootstrap consensus tree inferred from 500 replicates [16] is taken to represent the evolutionary history of the taxa analyzed. The evolutionary distances were computed using the Jukes-Cantor method [17] .
3. Results
This study has shown the frequency of Vibrio cholerae in different coastal communities and as shown in Table 1. Kaa community in Khana and Andoni both in Rivers South-East Senatorial zones showed prevalence rate of 16.0% and 30% respectively. For Ogu/bolo, out of 100 stool samples examined for Vibrio cholerae, 4 samples showed positive result, 16 samples had growth of other species of Vibrio (V. vulnificus and V. parahaemolyticus) indicating frequency of 4% while Abua/Odual (Rivers West) showed frequency of 10%.
3.1. Result of Antibiotic Susceptibility Testing
The total isolates from stool and sea food susceptibility to some selected antibiotics were calculated and represented in percentages (Table 2 & Table 3). The findings showed that the isolates from stool were susceptible to Ciprofloxacin and Gentamycin with susceptibility rate of 94.12% each while 100% resistance was recorded against Amoxicillin-clavulanic acid, 94.12% against Amikacin and 88.24% against Colistin. Hence, significant result was obtained. For the sea foods, the isolates were susceptible to gentamycin and ciprofloxacin with susceptibility rate of 91.43% and 82.86% respectively. Resistance was also recorded against Colistin (88.57%) and Azithromycin (82.86%).
3.2. Genomic Analysis
All of the isolates tested for presence of 16SrRNA gene were positive and show bands at 1500 bp. Plate 1 below showed representative of some bacterial isolates subjected to amplification for bacterial 16SrRNA gene.
![]()
Table 1. Prevalence of Cholera in different coastal communities in Rivers State.
![]()
Table 2. Antibiotic susceptibility testing for stool isolates.
X2 = 16.84, P = 0.032.
![]()
Table 3. Antibiotic susceptibility testing for seafood isolates.
X2 = 15.00, P = 0.09.
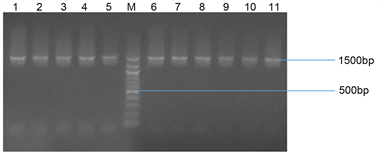
Plate 1. Agarose gel electrophoresis of some selected bacterial isolates. Lanes 1 - 11 represent 16 SrRNA gene bands (1500 bp). Lane M represents the 500 bp Molecular ladder.
3.3. Resistant Genes
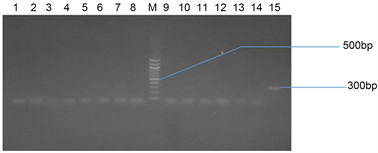
Plate 2(a) and Plate 2(b) represent the agarose gel electrophoresis of SHV gene of some selected bacterial isolates from seafood and stool samples respectively.
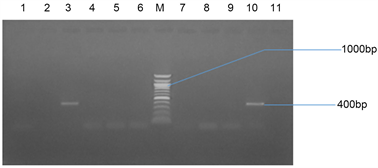
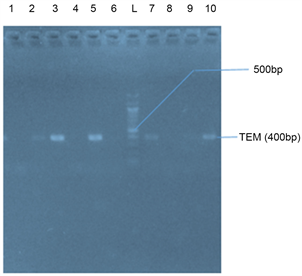
Plate 3(a) and Plate 3(b) represent the agarose gel electrophoresis of TEM gene of some selected bacterial isolates from seafood and stool samples respectively.
Plate 4(a) and Plate 4(b) represent the agarose gel electrophoresis of OXA gene of some selected bacterial isolates from seafood and stool samples respectively.
Plate 5(a) and Plate 5(b) represent the agarose gel electrophoresis of CTX-M gene of some selected bacterial isolates from seafood and stool samples respectively.

(a)
(b)
Plate 2. (a) Agarose gel electrophoresis of SHV gene of some selected bacterial isolates from seafood. Lane 1 - 11 represents a negative amplification of samples subjected to PCR. Expected base pair size of SHV (300 bp). Lane M represents the 300 bp Molecular ladder; (b) Agarose gel electrophoresis of some selected bacterial isolates from stool samples. Lanes 7, 10, 11 and 15 represent SHV gene bands (500 bp). Lane M represents the 500 bp Molecular ladder.
(a)
(b)
Plate 3. (a) Agarose gel electrophoresis of TEM gene of some selected bacteria isolates from seafood. Lanes 3 and 10 represent a TEM band (400 bp). Lane M represents the 1000 bp Molecular ladder; (b) Agarose gel electrophoresis of the amplified TEM gene from isolates of stool samples. Lanes 2, 3, 5, 7 and 10 represent the TEM bands at 400 bp while Lane L represents the 500 bp Molecular Ladder.
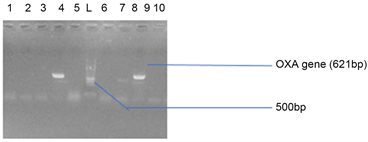
(a)
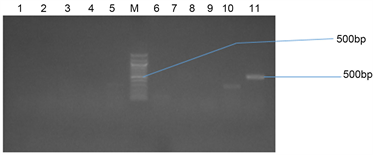
(b)
Plate 4. (a) Agarose gel electrophoresis showing the OXA bands in bacteria isolates from stool samples. Lanes 4 and 8 represent the OXA gene bands at 621 bp while lane L represents the 500 bp molecular ladder; (b) Agarose gel electrophoresis showing the OXA bands in bacteria isolates from stool samples. Lanes 11 represent the OXA gene bands at 500 bp while lane L represents the 500 bp molecular ladder.
3.4. Virulent Genes
Plate 6 and Plate 7 represent agarose gel electrophoresis showing the amplified AcrB and TDH gene bands.

(a)
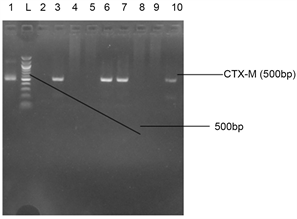
(b)
Plate 5. (a) Agarose gel electrophoresis showing CTX-M bands in some bacteria isolates from sea food samples. Lane 11 represents the CTX-M gene at 500 bp while lane L represents the 500 bp ladder; (b) Agarose gel electrophoresis showing CTX-M bands in some bacteria isolates from stool samples. Lane 1, 3, 6, 7, and 10 represent the CTX-M gene at 500 bp while lane L represents the 500 bp ladder.

Plate 6. Agarose gel electrophoresis showing the AcrB bands. Lanes 3, 6, 7, 8 and 9 represent the AcrB gene bands at 200 bp while lane L represents the 100 bp molecular ladder.
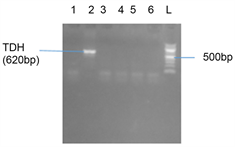
Plate 7. Agarose gel showing the amplified TDH gene band. Lane 2 showed the TDH gene band at 620 bp and Lane L represents the 500 bp ladder.
3.5. Phylogenetic Results for Genomics Study
Figure 1 shows the bacteria isolated from seafood that were identified from the gene bank and used to form the phylogenetic tree. The representative bacteria isolates from stool samples were also identified from gene bank and their evolutionary trend was demonstrated using Phylogenetic tree shown in Figure 2.
![]()
Figure 1. Evolutionary distances between the bacterial isolates from sea foods.
![]()
Figure 2. Evolutionary distances between the bacterial isolates from stool samples.
4. Discussion
In this study, the prevalence of cholera, antibiogram, genomics and phylogeny of bacteria from seafood and stool samples from some cholera-prone coastal communities in Rivers State were determined. The sample collection was distributed across the three Senatorial zones which include: Ngo Andoni, Kaa Khanan, Chukwuama Ogu/Bolo, Okrika, Finima Bonny and Egbolom Abua/Odual.
4.1. Antibiogram of Isolates from Stool Samples and Seafood
Antibiogram of isolates from seafood and stool samples to the commonly used antibiotics showed that the isolates expressed different levels of susceptibility and resistance to the antibiotics although the isolates identification using conventional procedures was not the same when compared with those identified using molecular methods, the discussion of the antibiogram pattern in this study was based on the molecular identification of the isolates. In the stool samples, Pseudomonas aeruginosa, Proteus mirabilis, Proteus vulgaris, Providencia stuartii, Enterobacter sichuanensis, Providencia sneebia and Klebsiella pneumoniae showed high resistance to Azithromycin, Colistin, Amikacin and Amoxicillin-clavulanic acid with percentage resistance of 88.24%, 94.12% and 100% respectively. All isolates apart from Klebsiella pneumoniae and Providencia sneebia showed resistance to Ceftazidime (64.71%). In general, the susceptibility rate of 94.12% towards Ciprofloxacin and Gentamycin was seen in the isolated bacteria including Klebsiella and Pseudomonas. There has been a report of increase resistance of Klebsiella to Gentamycin [18] in contrary to the finding of this study which had similar results with that of Nkemkanma et al. [19] . Considering the level of resistance of the isolates against Colistin in this study, the finding is not in agreement with the report of Bialvei and Samadi-Kafil [20] which reported resistance rate of less than 10% in Nigeria and stated that Colistin was returned to clinical practice to arrest the increasing lack of options for treatment of multidrug resistance Gram negative bacteria such as the ones isolated in this study. The result of Amoxicillin-clavulanic acid (AMC) is also in agreement with the results of Nkemkanma et al. [19] which also recorded 100% resistance against some bacteria such as Klebsiella pneumoniae. In this present study, all the isolates showed 100% resistance to AMC. Ciprofloxacin and Gentamycin showed better performance indices with each recording 94% susceptibility to all isolates and this agrees with finding of Roy et al. [21] . Isolates also showed high resistance to Amikacin. Amikacin is mostly used for treating severe infections with multidrug resistance aerobic bacteria especially Pseudomonas, Escherichia coli, Proteus, Klebsiella, Enterobacter, Serratia and Acinetobacter. Considering the level of resistance of some of these isolates to amikacin, it becomes worrisome as to the implication to Public health at large.
Similar isolates were obtained from seafood but most were susceptible to Gentamycin (91.43%) while high resistance rate was observed against Colistin (88.57%) and Azithromycin (82.86%). Microorganisms isolated from seafood include Providencia stuartii, Providencia rettgeri, Vibrio parahaemolyticus, Vibrio azureus, Aeromonas dhakensis, Klebsiella pneumoniae, Pseudomonas aeruginosa and Escherichia coli. High levels of antibiotic resistance recorded in this study could be as a result of excessive use of antibiotics in man [22] . More so, increase level of antibiotics is introduced into far m products and water in order to improve yield and treatment of infectious diseases [23] . The Vibrio parahaemolyticus isolated showed high susceptibility to cefotaxime and imipenem. This corroborates with the study carried out by Yang et al., [22] which also reported that examined Vibrio parahaemolyticus were susceptible to cefotaxime, cefoxitin, imipenem and meropenem. All the isolates showed multiple resistance to at least three different antibiotics even up to eight antibiotics. This ratio is greater than that reported in Xu et al. [24] . Vibrio azureus isolated from oyster exhibited over 60% resistance to antibiotics used in this study especially azithromycin, Colistin and Ertapenem.
4.2. Genes of Resistance and Virulence Present in the Isolated Bacteria from Seafood and Stool Samples
The common drugs of choice for treatment of severe infections caused by Gram negative bacteria belong to extended spectrum beta-lactam antibiotics [25] . Unfortunately, resistance to this group of antibiotics has emerged because of production of enzymes capable of hydrolysing beta-lactam family of antibiotics. These enzymes are termed extended spectrum beta-lactamases (ESBLs). They breakdown antibiotics such as Cephalosporins, Penicillins, Carbapenems, Monobactams and Cephamycins by hydrolysis of amide bond of beta-lactam ring and render the antibiotics inactive against their original cellular targets. They are inhibited by Clavulanic acids, Tazobactam and Sulbactam. In this study, isolates from seafood and stool samples that showed multidrug resistance were subjected to molecular detection of ESBLs genes (TEM, SHV, CTX-M and OXA). All the isolates from seafood except one (4%) did not express any of the resistance genes. Cefotaxime hydrolysing gene (CTX-M) was detected in Pseudomonas aeruginosa. The absence of resistance genes from multidrug resistance isolates obtained from seafood could be as a result of masked gene or gene sequence not similar to the sequences present in the gene bank [26] . For the isolates from stool samples, 5 (20%) showed CTX-M gene, 10 (40%) expressed TEM gene, 5 (20%) exhibited SHV gene while 2 (8%) showed OXA gene. The isolates harbouring both TEM and SHV genes were also identified (12%) as Proteus vulgaris, Escherichia coli and Klebsiella pneumoniae while isolates bearing both TEM and OXA and CTX-M and OXA were detected among 4% of the isolates each. The difference between results obtained from this study and those of other researchers have shown that ESBL gene may vary from one source of infection to another and from one location to another. This present study clearly showed the dramatic change in the gene pool of enterobacteriaceae found in coastal communities in Rivers State. The most prevalent bacteria bearing the resistance genes from this study were Escherichia coli, Enterobacter sichuanensis and Providencia sneebia. Similar studies have revealed Klebsiella pneumoniae and Escherichia coli as the major culprits [27] . The presence of ESBLs in Providencia sneebia and Enterobacter sichuanensis indicate that there could be spontaneous mutation or transfer of genetic material (plasmids) from one organism to another in a mixed culture. The presence of TEM gene in a higher number of isolates in this study than other resistance genes showed that this gene may be peculiar to particular regions with unique environmental activities. Similar study was carried out in Rivers state but from a different region and more of SHV gene was detected although the research did not consider TEM gene [27] . There are other reports of ESBLs in Nigeria implicating the enterobacteriaceae as carrier of ESBLs in clinical isolates [28] [29] . It was also observed that the OXA gene carrying isolates were resistance to Ertapenem and Amoxicillin-clavulanic acid and this is in agreement with the report of Bajpai et al. [26] .
In this present study also, the phenotypic examination of isolates against extended spectrum beta-lactam antibiotics showed higher resistance when compared with the actual expression of resistance gene as detected using genotypic method with 67.93% of multidrug resistance isolates not possessing genes TEM, CTX-M, SHV and OXA. It could be that those isolates that lacked the genes may have masked or hidden gene for ESBLs production. More so, genotypic procedure can only identify those genes with known sequences.
In addition to the detection of resistance genes from the isolates, presence of virulence genes were also assayed to ascertain the pathogenicity of the isolates especially from seafood since none of the isolates expressed the resistance genes. Pathogenicity is capability of microorganism to initiate disease condition. The pathogenic microbe possesses some attributes that enable to overcome host immunity and cause diseases. These attributes are collectively called virulence factors [30] . Most pathogenic microorganism combine their ability to produce toxin and also their ability to penetrate the host cell and spread to cause disease; but the complete infection process is dependent on the virulence of microorganism as well as host immunity. The evolution and distribution of resistance and virulence differ but despite their differences they share some similar features. Biologically, both processes are vital for microbes to survive adverse, unfavourable conditions because while virulence is important for microbes to evade host defence mechanism, the resistance to antimicrobials is very necessary to overcome antimicrobial therapy and survive in a competitive environment. Hence, immune defence mechanism and antimicrobial pressure are the logjams for survival of microbial community [31] . In this present study, two virulence genes were detected viz: the thermostable direct haemolysin (TDH) virulence gene associated with pathogenic Vibrio species and Efflux pump (AcrB) virulence gene associated with pathogenic enterobacteriaceae. These were assayed to confirm the pathogenicity of the isolated bacteria from stool and seafood samples used in this study. The TDH gene was detected in all the Vibrio parahaemolyticus isolated from oyster in Andoni community. This result coincides with the result of previous study which showed increase in Vibrio parahaemolyticus strain carrying TDH gene [32] . This gene was not detected in Vibrio azureus isolated also from oyster, indicating that either TDH gene is not associated with this specie of Vibrio or that this V. azureus is not pathogenic to man. The TDH production is strongly correlated with the pathogenicity of V. parahaemolyticus and expresses different biological features including haemolytic property, enterotoxicity and cytotoxic activity [22] . The V. parahaemolyticus isolated in this study showed multidrug resistance to Azithromycin, Cefotaxime, Ceftazidime and Colistin, indicating that this strain of Vibrio species has potential risk to public and environmental health. It was also observed that efflux pump gene was not detected in Vibrio parahaemolyticus. The reason could be that efflux pump gene may not be found in every bacterium.
There was high level of AcrB gene detected in both isolates from seafood and stool samples. In seafood, the AcrB was higher with over 50% increase than the prevalence in the isolates from stool samples. This could be as a result of resistance genes detected in isolates from stool samples. The gene AcrB encourages colonization, infection and persistence of enterobacteriaceae in the hosts and the intrinsic and acquired resistance to antibiotics are also enhanced by the efflux pump gene [33] .
When relationship between virulence and resistance was considered in this study, it was observed that isolates that expressed OXA resistance gene lack virulence genes. This is in agreement with the report of Beceiro et al. [30] which recorded decreased virulence gene in bacteria bearing resistance gene. This study also showed that resistance genes (SHV) and (CTX-M and OXA) were detected in Klebsiella pneumoniae and Enterobacter sichuanensis isolates respectively. These bacteria did not express any of the virulence genes assayed, supporting the observations [34] [35] [36] which reported that increase in antimicrobial resistance decreases the virulence of some Enterobacteriaceae. Beceiro et al. [30] had a different report about Klebsiella pneumoniae which possesses AcrB gene and was of opinion that AcrB is a key factor for both antibiotic resistance and virulence. In the Providencia sneebia isolated from stool samples, both resistance gene (TEM) and virulence gene (AcrB) were detected, indicating that their coexistence in the same bacterium will cause severe infection. This is in agreement with the findings of Beceiro et al. [30] .
4.3. Phylogeny of Isolated Bacteria from Seafood and Stool Sample
The obtained 16SrRNA sequence from the isolate produced an exact match during the mega blast search for highly similar sequences from the NCBI non-redundant nucleotide (nr/nt) database. The 16SrRNA of the isolate C1 showed a percentage similarity to other species at 100%. The evolutionary distances computed using the Jukes-Cantor method were in agreement with the phylogenetic placement of the 16SrRNA of the isolates from the sea foods within the Aeromonas, Vibrio, Pseudomonas, Escherichia, Klebsiella, Providencia and Proteus sp and revealed a closely relatedness to Aeromonas dhakensis, Vibrio azureus, Vibrio paraheamolyticus, Pseudomonas aeruginosa, Escherichia coli, Klebsiella pneomoniae, Providencia rettgeri, Providencia stuartii and Proteus mirabilis.
The phylogenetic placement of the 16S rRNA of the isolates from the stool samples were within Enterobacter, Pseudomonas, Escherichia, Klebsiella, Providencia and Proteus and revealed a closely relatedness to Enterobacter hormeachi, Enterobacter sichuanensis, Pseudomonas aeruginosa, Escherichia coli, Klebsiella pneumonie, Proteus vulgaris, Proteus mirabilis.
When the identified bacteria using conventional technique was compared with that of identified by 16SrRNA sequencing, it was observed that the isolates from the two identification approaches were different. The discrepancies so observed could be as a result of mutation or due to environmental factors affecting the use of culture media, cultural conditions and biochemical reagents. This finding is in agreement with Ollor et al. [37] which also opined that the difference was due to the fact that conventional techniques depend on the metabolic processes of microorganisms while molecular technique detect the unique deoxyribonucleic acid present in each microbe. The identified bacteria using molecular method could also cause clinical manifestation which would not have been discovered using conventional methods. Of all the bacteria identified using genomic technique, only Escherichia coli and Vibrio parahaemolyticus were identified using conventional method of isolation and identification.
5. Conclusions
From this study, it can be concluded that Vibrio cholerae is not the only bacterium responsible for cholera and associated gastrointestinal disorders. Other bacteria also exhibit clinical manifestations similar to that of cholera. Such bacteria include Enterobacter sichuanensis, Enterobacter hormaechei, Providencia sneebia, Providencia stuartii, Providencia rettgeri, Aeromonas dkakensis, Pseudomonas aeruginosa, Proteus mirabilis, Proteus vulgaris, Klebsiella pneumoniae and Escherichia coli. These bacteria were identified by sequencing the 16SrRNA from isolated bacteria. The study also demonstrated that change in weather condition influenced the presence of cholera in different coastal communities. This aspect of study has been published.
The seafood common to the inhabitants of the coastal communities was also studied to ascertain the relationship between the isolates from environmental samples and those from clinical samples and this study showed similar microorganisms were detected. The antibiogram of isolates from both stool and seafood was examined and all showed resistance to common antibiotics in use (Extended spectrum beta-lactam antibiotics and carbapenems). The isolates with higher resistance were examined for resistance genes and this study showed that most isolates from stool samples express ESBL resistance genes TEM, SHV, CTX-M and OXA. It was also observed that some isolates that showed resistance to antibiotics did not express any of the resistance gene. The virulence genes were also detected mostly from isolates gotten from seafood indicating that those isolates have higher potential to cause disease.
Declaration
Ethics Approval and Consent to Participate
All experimental procedures were approved by the Ethics Committee of Rivers State Hospitals Management Board, Nigeria.
Consent for Publication
Not applicable.
Availability of Data and Materials
The data presented in the study are included in the article.
Funding
The work is self-funded.
Author Contributions
A-CA, A-VN, N-EG and O-AO contributed to the study design, literature review, writing and compilation of manuscript. N-CU and W-CK revised it critically to meet publication standards. All authors read and approved the final manuscript.
Acknowledgements
Research Staff and Students of Diagnostic Laboratory Rivers State University Port Harcourt Nigeria, Prof Tatfeng MY of the Department of Medical Laboratory Science Niger Delta University, Amassoma Bayelsa State Nigeria.
Abbreviations
CTX-M: Cefotaxime hydrolysing capacity, CT: choleragen, TCP: toxin-Coregulated pilus, Omp: outer-membrane protein, PCR: polymerase chain reaction, DNA: Deoxyribonucleic acid, TCBS: Thiosulphate citrate bile salt, LB: Luria Bertani, NCBI: National Center for Biotechnology, Information, BLASTN: Basic Local Alignment Search Tool, Tgh: Thermostable Direct Haemolysin, ESBL: Extended spectrum beta lactamase, TEM: Temoniera, OXA: Oxacillinase, SHV: sulfhydryl variable, MEGA: Molecular Evolutionary Genetics Analysis, MAFFT: Multiple Alignment using, Fast Fourier Transform, Azm: Azithromycin, ctx: Cefatazime, CIP: ciprofloxacin, AK: Amikacin, CT: Colistin, CAZ: Ceftazidime, IPM: Imipenem, CN: Gentamycin, AMC: Amoxicillin.