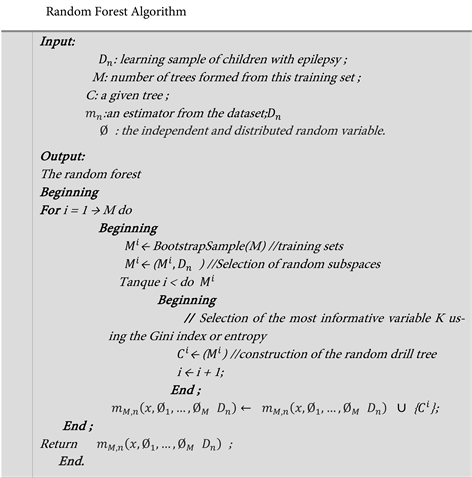
1. Introduction
Epilepsy is one of the most widespread and devastating neurological diseases in the world [1] . The hallmark of epilepsy is a set of abrupt and chronic changes in neuronal electrical activity causing recurrent seizures called “epileptic seizures” [2] . These misdiagnosed or ignored attacks can go unnoticed and intensify in the long term. Diagnosis of epileptic seizures is usually made by electroencephalography [3] . Electroencephalography is a technique that analyzes electrical signals from the brain. Those electrical signals can be represented as graphs called electroencephalograms (EEG). The EEG interpretation permits to detect the possible epilepsy presence. Nevertheless, for the patient to be fully cured without the disease sequelae in their mental health, it is necessary that the diagnosis be made at the disease onset. So, simpler and easier-to-use methods are needed for early diagnosis of the disease on a large scale. However, the visual interpretation of the EEG carried out by the epileptologist not only takes time, but can prove to be inefficient because of the disease visual identification mechanism complexity. Beyond these problems, the fact that seizures can be overestimated when the patient moves; which leads to noises and muscular artifacts disturbing the signal. Considering these circumstances, we wonder: How can we automatically detect epileptic seizures in newborns?
To address the aforementioned concern, we set the goal of proposing a machine learning model capable of efficiently identifying the presence or absence of epilepsy in the patient by essentially using electrical signals from the brain. Therefore, we have organized that article around three stages. The first part presents work on epilepsy; the second section deals with the methodology used. The third one presents the results of our work in which we obtained a random forest model for detecting neonatal epileptic seizures that is 99.4% correct.
2. State of the Art
The word epilepsy comes from the Greek “epilêpsia” [4] which designates a sudden attack, a sudden stop. That disease is considered a demonic disease thus preventing any scientific progress [5] . The negative prejudices surrounding the disease were obstacles to the proper care of patients, their development and social integration. It was at the beginning of the eighteenth century that Samuel Auguste David Tissot had looked into that very disease from a scientific approach in his book “Traité de l’épilepsie” in 1770 [6] . ESAMBERT, B. and ALLONNEAU-ROBERTIE, E. proved that 20% of children with epilepsy showed a significant backwardness in certain learning areas, in particular reading and sport [7] . According to the work of Vibha Patel et al. [8] [9] , the method of detecting epileptic seizures can be classified into four categories. It is a method based on the traditional machine learning approach, the deep learning approach, Isignal processing approach and hybrid approach. Artur Gramacki and Jarosław Gramacki, on the other hand, focus on the deep learning method. Although the said approach presents a very good performance, but it is all the same non-parametric and requires a large number of data for its configuration [10] . P. Geethanjali on the other hand, asserts that no method has so far shown both high sensitivity and zero false alarms to obtain reliable results [11] . P. Fergus, D. Hignett, A. Hussain, D. Al-Jumeily, K. Abdel-Aziz [12] [13] automatic detection of epileptic seizures using EEG [14] [15] . Using the aforementioned tool, the authors were able to extract the characteristics of a person living with epilepsy via their scalp. C. Donos, M. Dümpelmann, A. Schulze-Bonhage [16] also used the intracranial EEG tool to detect seizures early. The authors use the same characteristics which gave them an average sensitivity of 93.84%. Likewise Jerry Yu [17] used the EEG to classify epileptic seizures. The method enabled them to classify the disease as epilepsy with an accuracy of 84.2%.
However, rare are the studies which are interested in the issue of epilepsy in newborns. Hence the interest of proposing a model capable of evaluating the presence or absence of that disease in children under one year of age.
3. Methodological Approach
As part of the study we used the Electrophysiological technique. That technique is based on the EEG (Electroencephalography) method and uses electrodes placed on the surface of the head to collect recordings from the cerebral cortex via the EEGLab software [15] . The data comes from epileptic children and listed in Figure 1 below. We were able to obtain ten (10) EEG files that were preprocessed from the EEGlab software, labeled using the annotations of the three (3) experts, then concatenated. The new database consists of 1048544 seconds of annotated lines of recordings including the variables that are the EEG signal sensors placed on the head of infants.
The input variables in table 1 above are sensors of the EEG signals which are: Fp2, F4, C4, P4, O2, Fp1, F3, C3, P3, O1, F8, T4, T6, F7, T3, T5, Fz, Cz, Pz as well as the ECG/EKG and Resp-effort which collect the amplitudes of the cerebral, cardiac and respiratory activity of the patients. The designations of the sensors represent the positions at which they are placed on the patient’s skull such as: F stands for Frontal, P stands for Parietal, T stands for Temporal, O stands for Occipital, C: Central and z represented the middle line. The even numbers 2, 4, 6, 8 represent the right hemisphere while the odd numbers 1, 3, 5, 7 represent the left hemisphere. The ECG/EKG corresponds to the electrocardiogram and Resp-effort corresponds to the infant’s breathing effort. Our response variable y constitutes the signal state at that second (epileptics or non-epileptics).
4. Modelization
Through the present article, we evaluate the entropy measure and the Gini index in the detection of epilepsy disease in newborns. The Gini index allows to measure
Figure 1. Text file resulting from the extraction of the selected components.
the membership frequency of a random element in a subset. While entropy is a measure of disorder in a data set and it is used to choose the value to maximize information gain. So, x let us consider the explanatory variables vector of the epileptic children as follows:
of the explanatory variables is with values in
. Let Y us set the random variable of children with epilepsy to be predicted. Consider
the set of n-samples of children with epilepsy as follows:
independent and identically distributed random variables with the same distribution as the couple
. Let M us pose a collection of random trees and the estimator associated with these M trees. For the iième forest tree, the predicted value at a new x point is noted:
with
they are independent random variables, variable
distributed and independent of
. Thus, the random forest estimator, which results from the aggregation of the different trees is given by the following formula:
5. Simulation and Results
The programming of the neonatal epileptic seizure detection algorithm was done using the python programming language in the Jupyter programming interface with the Scikit-Learn library. The simulation was carried out on the data in Figure 2 below.
This database contains 1,048,544 observations and 23 variables that we will have subdivided into two different sets. The first is for training 70% and another set for validation 30%. All training of the models contains the largest number of data which is 733,981 therefore the validation set contains the remaining 314,563 data. After sampling, we trained four (4) models. During the learning we vary the hyperparameters of the models as shown in Figure 3 below:
![]()
Figure 2. Database of children with epilepsy.
We observe in Figure 3, the results of our models on the training sample to which we applied a validation 5-fold cross validation assessed by accuracy metric.
Following the training of our machine learning model of supervised classification using our learning sample, we observe that the optimized models obtained have very high performance. Despite the extreme differences in EEG readings between patients having a seizure and not having one. Moreover, by observing the accuracy index, we can attest that they distinguish very well between our two classes which are: presence of epileptic seizure/absence of epileptic seizure. However, we make the following observation:
The variant accuracy of the random forest model using entropy and a number of trees; 100 is 0.986; which is higher than the accuracy of other optimized models. We can then conclude that our best model is the random forest model (100; entropy). We then applied our validation data to our best model and obtained the following performance metrics as results:
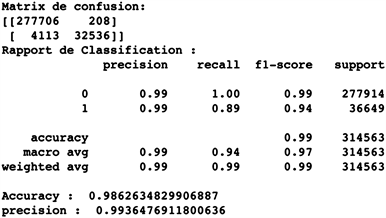
An accuracy of 0.986; a precision of 0.994 and a recall/specificity of 0.994.
6. Results Interpretation
Our random forests model scores very high on the accuracy, recall, and precision metrics with the test set. With an accuracy of 0.986; an accuracy of 0.994 and a recall of 0.994 in the test, our random forests classifier is a very powerful model.
To corroborate it, we observe the predictions of the confusion matrix in Figure 4 below, we have:
277,681 true negatives versus 188 false negatives;
32,464 true positives versus 4230 false positives.
It gives us an error of about 1.4% on the validation dataset and means that the model excels at the difference between the two (2) classes (positive and negative samples).
We have created a machine learning model of neonatal epileptic seizure detection based on the Random forests classifier algorithm that can predict whether patients are having a seizure or not through EEG readings. It is correct to 99.4% to predict positive classes in the test set. The following Figure 5 shows us the performance of the learning accuracy and the validation accuracy:
![]()
Figure 5. Representation of model performance.
We observed that the two curves are globally increasing. And it means that the model presents a large ability to learn the data characteristics in order to make effective predictions. Also, the small distance between the peaks of the two Accuracy curves demonstrates that the model is neither overfitting nor under-fitting. The model has good generalizability to other data.
The more the model will be used on new data, the more its accuracy will be increased and we will observe a low rate of false prediction.
7. Conclusion
The article presented a model to detect neonatal epileptic seizures. It is a supervised machine learning model combining the extra trees classifier with 5-fold cross validation for the implementation of a very efficient model. The present work allowed us to develop an automatic epilepsy crisis detection model from the electroencephalogram signals amplitudes in human newborns. To realize that work, the EEGlab software was used to preprocess and segment the EEG signals of 10 newborns. And the python programming language allowed us to implement the model. The resulting model, based on the extra trees approach, is able to predict the existence or absence of epilepsy with an accuracy of 99.4%. The use of the said model will greatly reduce the waiting time for the results of examinations and thus improve the time taken to take care of patients.
Acknowledgements
The authors would like to thank the Virtual University of Côte d’Ivoire, the Ivorian Ministry of Health and Dr. ACHIEPO Odilon Yapo Melaine for their contribution to the success of this research work.