Repurposing Antidiabetic Drugs against Respiratory Syncytial Viral Infection: A Docking Study ()
1. Introduction
Respiratory Syncytial Viral Infection is one of the most common viral infections in the respiratory airways and is more common in infants, younger children and some adults [1]. After viral entry into human, they can cause severe pneumonia, bronchitis and can result into hospitalization. Death resulting from this infection has also been reported in developing countries [2]. This necessitates the research into drug design and discovery to curtail this menace. Although Ribavirin, an indirect inhibitor of RNA transcription has been used in the management of the RSV infections, a lot of molecules are still under development with the overall goal of improving the quality of life of the populace [3]. N4-hydroxycytidine has been reported to be a potent inhibitor of the RSV infection but 36% to 56% dose dependent oral bioavailability was demonstrated [4].
RSV is an enveloped single strand RNA Virus with two enveloped glycoprotein G and F which are crucial for the viral attachment and entry into the host cell membrane [5]. Once the virus binds to the receptor of the host cell via its G protein, it induces a conformational change on its F protein and allows the Fusion Peptide insertion into the host cell membrane, bringing the interaction of three HRA domain and three HRB domain together and consequentially forming a six helix bundle fusion core [6] [7] [8]. This finally brings the cell membrane and the viral membrane in close proximity to each other for fusion and viral entry [9].
Structure guided drug design approach has been very useful in the development of effective antiviral agents from insilico studies to experimental validations and clinical trials based on the insight from the pathogenesis and mechanism of viral entry [10]. Also, plants with ethno medicinal claims such as antitussive, antiviral, antibacterial, antidiabetic properties have been reported and have been validated with bioassay [11]. Quercetin and Quercetin pentaacetate, flavonoid from plant block the viral adhesion via insilico and in vivo model. Although some researches criticize the poor solubility of Quercetin, hence the need is for its acetylation [12].
Challenges in terms of pharmacokinetics properties, side effects and route of drug administration optimization of existing lead molecules demand for a research for an effective drug with good ADME properties.
Herein, we repurpose a class of pharmacological agents with an attempt of interrupting with the formation and or stability and the six helix bundle fusion core using insilico approach. 14 FDA antidiabetic drugs were screen against the F protein of the RSV. Four of them are identified as drug candidate for further experimental validation and optimization.
2. Materials and Method
2.1. Data Sources
14 FDA Approved antidiabetic drugs were obtained from online sources and there chemical structures were retrieved from Pubcem.
2.2. Preparation of Target
Cocrystallized ligand with the PDB ID:3KPE and resolution of 1.47 Å was retrieved from the protein data bank database; RCSB PDB [13]. Before the docking process, charges were assigned, water of crystallization were removed and the protein was further optimized by the Autodock tool and Biovia Discovery Studio Visualizer before the docking process [14].
2.3. Preparation of Ligand
The 3D SDF structure of all the 14 compounds was downloaded from Pubchem database. Following automated energy minimization and optimization of all the ligands [15], they were all converted to PDBQT format using the graphical user interface version of PyRx virtual screening tool-python prescription 0.8.
2.4. Compound Screening Using PyRx Program
Virtual screening of all the compounds was done using PyRx software by Autodock wizard as the engine for docking [16] [17]. The configuration file for the grid parameters was generated using Auto Grid engine in Pyrex. Predefined XYZ Center Coordinate of 23.782981, −7.656667, −2.076111 respectively having amino acids in the active site of the protein were set using the Vina wizard. The results less than 1.0 Å in positional root-mean-square deviation (RMSD) were considered ideal and clustered together for finding the favorable binding. The ligands with the highest binding energy were considered to be ligands with very high affinity.
Analysis of Result: The result were analyzed using Biovia discovery studio 2021 visualizer program and the receptor-ligand interactions of the various conformations were studied in their 2D and 3D format.
3. Result and Discussion
Respiratory Syncytial Viral is one of the most common form viral infection in infants and children. A druggable target such as the formation of the Six Helix bundle core has been identified and molecular docking has been used as a template in drug development. Molecular docking studies about the non-covalent interactions between ligands and target receptor site and provides there binding affinity as shown in Table 1. It also provides the preferred orientation of a molecule and suggests a molecule with a good binding energy as a good drug candidate. Our study screened 14 antidiabetic agents against the formation of the Six Helix Bundle fusion core; which is crucial for the viral fusion and entry. HR1 (Y198) and HR2 (D486) have been reported to play a crucial role in viral binding and entry [18]. Interestingly, Four (4) of the ligands showed a high binding affinity at the active site which are in the order of Glibenclamide (−7.7), Glimeperide (−7.7). Glipizide (−7.6). Canagloflozin (−7.5). They either interrupt the six helix bundle formation or the stability of the helix bundle and will be a good drug candidates for experimental validation. In terms of the classes of the drug, sulfonylurea family showed a great lead compared to other classes and the control. The four promising ligands bind to (TYR 198, ILE 492, ASP 489), (ILE B: 492, PHE B: 483, TYR A: 198, ASP A: 194, ASP B: 486 LYS A: 201), (GLU B: 487, SER B: 485, ASP B: 486, TYR A: 198, GLN A: 202, LEU A: 195 PHE B: 488 ASP A: 194 ASN A: 197, LYS A: 201), (LYS A 201, TYR A: 198, ASP B: 489, ASP B: 486, PHE B: 488) respectively as shown in Figures 1-5.
![]()
Table 1. Oral hypoglycemic agents and their binding Energy with the helix bundle of the Respiratory Syncytial Virus.
![]()
Figure 1. 3D interaction of glibenclamide at the active site.
![]()
Figure 2. 2D interaction of Glimepiride at the active site.
![]()
Figure 3. 2D interaction of Glipizide at the active site.
![]()
Figure 4. 2D interaction of Canagliflozin at the active site.
![]()
Figure 5. Canagloflozin orientation at the active site.
One of the advantages of repurposing this class of drugs is that in terms of subjecting them to clinical trials after the experimental validation, they will not be time consuming as regards their pharmacokinetic optimization because they have been used in clinical settings before now and they conform to the Lipinski rule of Five [19]. Furthermore, these ligands can be considered as prophylactics in infants and adults via the inhalation route.
TWO DIMENSIONAL STRUCTURES OF THE VARIOUS ANTIDIABETIC AGENTS
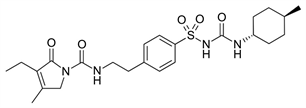
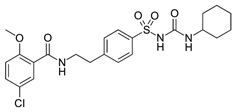
A B
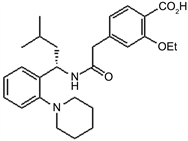
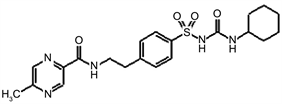
C D
E F
G H
I J
K L
![]()
![]()
M N
![]()
O
4. Conclusion
Drug repurposing will be the best approach to develop effective therapeutics against the Respiratory Syncytial Virus. Although ribavirin has been used in the management of RSV infection, many effective drugs are still under development. In this study, we have used Pyrex, Autodock tools and Biovia discovery studio tool to identify potential inhibitors of the formation of the six helix bond formation and/or stability; a pathway crucial for RSV viral fusion and entry. We screened 15 ligands and 4 of them are potential drug candidate for experimental validation, preclinical trials, clinical trials and route of administration optimization. To the best of our knowledge, this is the first time of this drug repurposing study and the identification of lead molecules against RSV from oral hypoglycemic agents.