In this work empirical models describing
sampling error (
Δ) are reported based upon analytical findings elicited from 3
common probability density functions (
PDF):
the Gaussian, representing any real-valued, randomly changing variable
x of mean
μ and standard deviation
σ; the Poisson, representing counting data:
i.e., any integral-valued entity’s count
of
x (cells, clumps of cells or
colony forming units, molecules, mutations, etc.) per tested volume, area, length of time, etc. with population mean
of
μ and
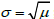
; binomial data representing the number of successful
occurrences of something (
x+) out of
n observations or sub-samplings. These data were generated in such a way as to
simulate what should be observed in practice but avoid other forms of
experimental error. Based upon analyses
of 10
4 Δ measurements, we show that the average
Δ (

) is proportional to
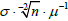
(
σx•μ-1; Gaussian) or
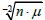
(Poisson &
binomial). The average proportionality constants associated with these
disparate populations were also nearly identical (
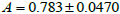
; ±
s). However,
since
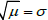
for any Poisson process,
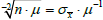
. In a similar vein, we have empirically demonstrated that
binomial-associated

were also proportional to σ
x•μ
-1. Furthermore,
we established that, when all

were plotted against
either
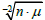
or σ
x•μ
-1, there was only one relationship with a slope =
A (0.767 ± 0.0990) and a near-zero
intercept. This latter finding also argues that all

, regardless of parent
PDF,
are proportional to σ
x•μ
-1 which is the coefficient of variation for a population
of sample means (
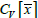
). Lastly, we establish that the proportionality constant
A is equivalent to the coefficient of
variation associated with
Δ (
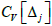
) measurement and, therefore,
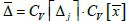
. These results are noteworthy inasmuch as they provide a
straightforward empirical link between stochastic sampling error and the
aforementioned
Cvs. Finally, we demonstrate that all attendant empirical
measures of
Δ are reasonably small (e.g.,
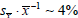
) when an environmental microbiome was well-sampled:
n = 16 - 18 observations with μ∼3 isolates per
observation. These colony counting results were supported by the fact that the
two major isolates’ relative abundance was reproducible in the four most
probable composition observations from one common population.