1. INTRODUCTION
Protein design should retain both the folding efficiency and functionality of an enzyme [1]. In nature, a protein’s sequence and interactions between residues in the protein core are not fully optimized and only achieve the minimum requirements for proper functioning. This situation leaves plenty of room for improvement [2,3]. Increasing the hydrophobic effect between residues in the protein core, the stability of the protein could be further enhanced [4,5]. Protein stabilities have been described to increase linearly with increasing hydrophobicity of the substituted residues [6]. However, residue substitution in proteins could result in numerous uncertainties. Hence, protein prediction softwares were used to simplify folding pathways and anticipate efficient, as well as inefficient mutations.
T1 lipase from Geobacillus zalihae was chosen for this study as it has been well studied and the structure is available [7-9]. The lipase shares high sequence similarity with the BTL2 lipase from Bacillus thermocanulatus. The oxyanion hole of T1 lipase was inferred as residues Q114 and F16, based on the open-form crystal structure of the BTL2 lipase [10]. We believed that the role of hydrophilic Q114 was not just limited to the binding with the substrate. The residue also regulated conformational stability and conferred specific enzyme attributes to T1 lipase. Since Q114 is positioned deep in the catalytic pocket, its substitution could alter protein compactness in the vicinity of the mutation and affect conformational stability. The close proximity of Q114 to the catalytic machinery also increased the prospect of transforming the shape of the catalytic pocket. This could lead to alternative enzyme-substrate interactions which are manifested as enzyme attributes that are very different from the wild-type.
There have been no reports on how the mutations at the oxyanion Q114 could affect T1 lipase. In-silico mutagenesis was performed to anticipate the possible physical changes resulting from the mutations. Then, we substituted the hydrophilic Q114 with six other hydrophobic residues, namely, Phe, Met, Ile, Leu, Val and Trp, respectively. The purpose of this article was to demonstrate that by varying the hydrophobicity of the oxyanion at site 114, the conformational stability and enzyme attributes of T1 lipase protein could be modulated. The stability and attributes of the six lipase variants were evaluated under variable hydrolysis conditions and compared to the wild-type. The residues which positively and negatively affected T1 lipase were identified.
2. EXPERIMENTAL
2.1. Materials
The growth media components were procured from Difco Laboratories (USA). The Quick-ChangeTM SiteDirected Mutagenesis Kit was purchased from Stratagene (La Jolla, USA). Agar plates were prepared by the addition of tributyrin to Luria-Bertani (LB) agar (Oxoid, UK) and antibiotics, Bradford reagent, chloramphenicol (35 mg/mL) and ampicillin (50 mg/mL) were supplied by Amresco (Ohio, USA). The culture harboring the recombinant T1 lipase gene was obtained from stock cultures from the Enzyme and Microbial Technology Laboratory, UPM. The BL21 (DE3) pLysS cultures were from Invitrogen (Groningen, Netherlands). Electrophoresis grade acrylamide, sodium dodecyl sulfate and calcium chloride dehydrate were purchased from Sigma-Aldrich, USA. The various surfactants were purchased from Merck (Germany). Glutathione-Sepharose HP and Sephadex G250 resin was procured from GE Healthcare (USA). Corn oil, soy oil and sunflower oil were acquired from Mazola, Malaysia. Coconut oil was obtained from Parachuta, Malaysia. Rice bran oil was purchased from Pura Harvest, Malaysia. Olive oil was procured from Bertolli, Italy. The solvents used were of HPLC grade and were obtained from Sigma-Aldrich Inc. (St. Louis, MO, USA). Membrane filter was from Sartorius (Germany) and the Amicon Ultra-15 centrifugal filter was from Millipore (Bedford, USA). Metal ions, calcium chloride, potassium chloride, lithium chloride, manganese chloride, ferrous chloride, strontium chloride and zinc chloride were purchased from Fluka, Switzerland. Gum Arabic, potassium hydrogen phosphate and potassium hydrogen orthophosphate, pyridine and magnesium chloride were procured from Ajax, Australia. Glycerol, 99% was obtained from BDH, UK. Deionized water was produced in our laboratory.
2.2. In-Silico Mutation and Estimation of Protein Structural Changes
FoldX software, version Beta 3.0, was used for the insilico mutation and estimation of the protein stability of the crystal structure of T1 lipase (PDB code: 2DSN). Residues with bad torsion angles were identified and repaired. Six mutants were prepared by substituting Q114 with Val, Leu, Met, Ile, Trp and Phe, respectively. Possible conformations of the substituted amino acids were analyzed and subjected to simple energy minimization to remove high-energy local minima, followed by the calculation of protein stability. The protein packing of each lipase variant was further analyzed with FoldX to evaluate how compact each protein was and to estimate the number of total cavities. The three-dimensional structures of the lipase variants built in FoldX were submitted to the standalone Voronoia 1.0 program.
Voronoia 1.0 software was used to estimate the atomic packing density (PD) and number of cavities [11]. The calculation is as shown below in Equation (1):
 (1)
(1)
whereby, PD refers to the atomic packing density of a particular protein, V(VdW) is the van der Waals volume of an atom, referring to the space inside the atom’s van der Waals radius that has been extended by 1.4Ǻ and V(SE) refers to solvent excluded volume. Both, the atoms’ van der Waals Volume and the Solvent Excluded Volume are cut by separating surfaces with atoms in very close proximity. In a perfectly packed structure, the V(SE) would be occupied entirely by separating surfaces and the packing density value is 1.0.
The cavities present in the protein are defined by Voronoia 1.0 as packing defects large enough to enclose at least one water molecule. The cavities are totally buried locations to which a 1.4Ǻ virtual atom could be placed. The probe also should not intersect any atoms’ van der Waals sphere and the cavity must not extend to the protein surface. Based on the assumptions, the cavities were estimated accordingly [12]. The program is a free database downloaded from the website http://bioinformatics.charite.de/voronoia.
2.3. Substitution of Residues by Site-Directed Mutagenesis
The same six mutant proteins described above were made using the QuickChange method. The PCR reaction mixture was prepared according to protocol provided by the manufacturer. The primers were designed so that sequence change was positioned in the center of the primer with approximately 10 - 15 bases on both sides. The DNA was incubated with the restriction enzyme Dpn-1 at 37˚C for 1 h to digest the parental methylated DNA. The Dpn-1-digested DNA was introduced into competent E. coli BL21 (DE3) pLysS cells, and the cultures were grown on LB-tributyrin agar media supplemented with ampicillin and chloramphenicol. Each mutation was confirmed by sequencing. Glycerol stocks were then prepared and kept at –80˚C.
2.4. Partial Purification of Enzymes
The recombinant culture (2000 mL) was harvested by centrifugation and re-suspended with 40 ml of PBS (pH 7.4) containing 5 mM of dithioreitol prior to sonication. The cell lysate was cleared by centrifugation at 12,000 g for 30 min and filtered with 0.45 µm membrane filter. Glutathione-Sepharose HP resin (10 mL) was packed into XK 16/20 column and was equilibrated with tencolumn volume of PBS (pH 7.4). The cleared cell lysate was loaded on a Glutathione-Sepharose HP column at a flow rate of 0.25 mL/min. The column was washed with the same buffer until no protein was detected. The bound lipase was eluted with Tris-HCl buffer, 100 mM NaCl and 0.33 mM CaCl2, pH 8.0. The active fractions were determined by SDS-PAGE gel electrophoresis and pooled, followed by concentration using Amicon Ultra- 15 centrifugal filter. The concentrated solution was subjected to gel filtration in Sephadex G250 in a XK16/20 column and pre-equilibrated with PBS buffer pH 7.4. The solution was run in the same buffer at flow rate of 1 mL/min. The active fractions were collected and concentrated with Amicon Ultra-15 centrifugal unit. The homogeneity of the partially purified protein was confirmed by SDS-polyacrylamide gel electrophoresis.
2.5. Standard Lipase Assay and Protein Concentration
The method of Kwon and Rhee (1986) was used to determine the lipase activity of the variants [13]. The culture filtrate of 1 mL (30 uL enzyme in 970 uL 50 mM glycine-NaOH buffer pH 9.0) was shaken with 2.5 mL of substrates: oil emulsion (oil: 50 mM glycine-NaOH buffer (1:1, v/v), with gum Arabic (0.0001%, w/v) and 20 μL of 0.02 M CaCl2 in a water bath shaker at 200 rpm. Each emulsion was prepared by mixing together an equal volume of olive oil (Bertolli, Italy) with 50 mM glycineNaOH buffer pH 9.0 and gum Arabic (0.0001% w/v) and the mixture was vigorously stirred for 10 min. The reaction mixture was shaken for 30 min at appropriate temperatures and terminated by adding 1 mL of 6 M HCl and 5 mL isooctane, followed by vortexing for 30 s. The upper isooctane layer, a volume of 4 mL containing the fatty acid was transferred to a test tube containing 1 mL copper reagent and vortexed again for 30 sec. The reagent was prepared by adjusting the copper(II) acetate-1- hydrate solution 5% (w/v) to pH 6.1 with pyridine. Lipase activity was determined by measuring the amount of free fatty acid released, based on the standard curve of oleic acid. The production of free fatty acids was monitored at 715 nm in a spectrophotometer (HITACHI U- 3210) with isooctane as blank. One unit of lipase activity is defined as the amount of enzyme releasing 1 μmole of fatty acid/min.
Finally, the standard assay for the determination protein content was carried out according to the method of Bradford (1976), using a standard curve prepared from standard solutions of bovine serum albumin (BSA: Sigma, USA) and Amresco assay reagent. The protein content was monitored at 595 nm in a spectrophotometer (HITACHI U-3210) using solution without bovine serum albumin as blank [14].
2.6. Lipase Characterization
The following standard lipase assay for the characterization of the lipase variants were performed in triplicates and in two determinants. The results were the mean of the obtained experimental reading. The specific activity of each lipase variant was compared to each other and the wild-type. Specific activity is the amount of substrate converted by the lipase per unit of time as per mg protein in the sample.
The effect of the substitution on the optimum temperature of the lipase variants were evaluated at various temperatures ranging from 40˚C to 80˚C under agitation rate of 200 rpm and incubated for 30 min. The substrates preference test was performed using natural oils (olive oil, soybean oil, corn oil, sunflower oil, rice bran oil and coconut oil) emulsions in the standard lipase assay. The lipase activity in organic solvents of different log P was also evaluated. The solvents were added to the enzyme at a ratio of 1:3 and incubated for 30 min at 30˚C. The effect of various surfactants on the lipase activity was carried out with surfactants added directly to the lipase assay at final concentration of 0.1% v/v. The effect of various metal ions was evaluated by direct addition of the metal ions into the standard assay. The metal ions, Li(I), Na(I), K(I), Ca(II), Mg(II), Sr(II), Fe(III) and Zn(II) were added to achieve a final concentration of 1 mM.
3. RESULTS AND DISCUSSIONS
3.1. In-Silico Mutation and Estimation of Protein Structural Changes
Table 1 depicts results from the in-silico studies using Voronoia 1.0 and FoldX. According to FoldX, the stability of the lipase variants improved over the wild-type when the hydrophobicity of the substituting residue was increased, with the exception of the Phe and Trp lipase variants. Replacements of Phe and Trp into Q114 were expected to adversely affect conformational stability of the variants, with the Trp residue being most destabilizing. FoldX predicted, in descending order, that the Met,

Table 1. Characteristics of the lipase variants and wild-type, T1 lipase acquired from in-silico studies using FoldX and Voronoia 1.0.
Leu, Ile and Val mutants will be more stable than the wild-type. The lower values of the calculated total energies in the variants could be correlated to the more hydrophobic nature of these residues.
On the other hand, analysis by Voronoia 1.0 showed both, mixed and inferior protein qualities. Interestingly, Voronoia 1.0 anticipated the Phe variant to be more stable due to a higher packing density. However, the one unit increase in the number of cavities in the Phe variant, nullified the benefit of a higher packing density and could lead to a less stable protein structure. This was corroborated by results in FoldX which predicted an increase in the total energy of the Phe variant, suggestive of a less stable lipase protein. Meanwhile, Voronoia 1.0 also estimated that substitution of residues Val, Leu and Met could result in cumulative inferior protein qualities. The less stable protein structures were exemplified with increased number of cavities and lower protein densities in the Val, Leu and Met variants. Voronoia 1.0 predicted only the Ile variant to have comparable stability as the wild-type.
A less dense atomic packing in Val, Ile, Leu and Met variants estimated by Voronoia 1.0, suggested that the internal amino acids within the catalytic pocket were more loosely packed. This caused the enzyme molecule to slightly increase in size and negatively affected the structure of T1 lipase. However, the lower total energy values predicted by FoldX for the Val, Ile, Leu and Met variants implied that mutation at Q114 also positively modified the internal structure and improved their catalytic attributes. It was concluded that placement of hydrophobic amino acids at site 114 had both, positive and negative impact on the structure of T1 lipase. The conformational stability and enzyme properties of T1 lipase were also affected.
3.2. Optimum Temperature
Figure 1 denotes the temperature-activity plots for determination of the optimum temperature of the lipase variants. The Val, Met and Trp variants exhibited lower optimum temperature at 60˚C. Substituting Q114 with Ile and Phe residues afforded variants that retained the optimum temperature as the wild-type at 70˚C. Meanwhile, the Leu substituted variant showed a remarkable increase in optimum temperature to 80˚C.
The significantly higher optimum temperature in the Leu variant could probably be attributed to an improved protein packing as a result of the smaller Leu side chain. Furthermore, when compared to Gln, substitution with the more hydrophobic Leu would have increased hydrophobic interaction between adjacent hydrophobic amino acids within the vicinity of the mutation. According to Ishikawa et al. (1993), introduction of a methylene group into the protein core can enhance protein stability through the increase in hydrophobic interactions [15]. Presence of Leu may have facilitated local structure folding when the bulky hydrophilic Gln was replaced with the hydrophobic Leu residue [16]. Moreover, the probability of conformational change with resulting deletrious effects was much lower when Leu was substituted into Q114. Hydrophobic amino acids have intermediate levels of side-chain flexibility [17]. Replacement with Leu could result in a more compact and rigid structure that was less susceptible to high temperature deactivation. Readjustment of the protein core to its size, presumably increased the overall compactness and tends to restore protein stability [18].
On the other hand, the hydrophilic Q114 in the wildtype was likely to cause more flexibility within the catalytic pocket. It has been reported that the inherent flexibility of amino acid side-chains, independent of backbone influences, could contribute substantially to the apparent flexibility in the surrounding area of the mutation point. Large polar or charged residues such as Glu, Gln, Arg and Lys, as well as serine, have been proven to be more flexible, while the aromatic amino acids, were

Figure 1. The activity plots as a function of temperature in the investigation of the optimum temperature of the lipase variants, Phe, Met, Ile, Leu, Val, Trp and wild-type T1 lipase for hydrolysis of olive oil. The reaction mixtures were assayed for 30 min with agitation at 200 rpm. Each temperature investigation was prepared in triplicates in two determinants and the results were the mean of the obtained experimental reading.
less so. Previous investigations have described increased amino acid flexibility near the mutation sites following substitution with these residues [17]. The Q114 in the wild-type would have increased the propensity towards temperature-induced conformational change due to a more flexible protein structure [19], particularly near the active site. This was reflected in the enzyme’s lower optimum temperature as opposed to the Leu variant.
Generally, proteins are well optimized in their structures for their physiological conditions and, thus, it is reasonable that the T1 lipase from Geobacillus zalihae is inherently thermostable. Nevertheless, modifying the residue at location 114 to at least six other residues, could still further enhance the activity of the lipase. This indicated that this location has not been fully optimized with respect to thermal activity in the course of evolution.
3.3. Substrates Preference
In this investigation, substituting the oxyanion Q114 with six different hydrophobic amino acids afforded mixed results. The Met and Trp substituted variants were more efficient than the wild-type in hydrolyzing the various natural oils as depicted in Figure 2. The highest activity was noted in the Met variant for all of the tested oils except in olive oil. The wild-type preferentially hydrolyzed corn oil and sunflower oil followed by rice bran oil, soybean oil, olive oil and coconut oil. However, substitution with Met at Q114 changed the preference to olive oil.
There was also marginal improvement in the Trp variant over the wild-type for the hydrolysis of olive oil and corn oil, but corn oil was hydrolyzed preferentially by the Trp variant. The Leu variant exhibited almost comparable lipase activity, except in corn oil and sunflower oil. Replacement of Q114 with Leu and Ile favored efficient hydrolysis of saturated, short-chain coconut oil. A higher hydrolysis rate was observed for sunflower and rice bran oil when Q114 was mutated to Phe. However, a significant decline in function was observed for the Val, Ile and Phe variants. The reduction was more pronounced in lipases substituted with Val and Ile as their catalytic activities were reduced to less than 30% of the wild-type. The former preferred soybean oil while the latter was partial towards coconut oil.
The enhancement in hydrolysis of natural oils when substituting Met into Q114 suggested that the Met residue might have altered interactions between the enzyme and substrate. The Met residue could have eliminated the hydrogen bond between the oxyanion Gln side-chain and the carboxyl group of the triglyceride [20]. This could partially explained the higher specific activity and improved hydrolysis of the natural oils by the Met variant over the wild-type. Moreover, the observed differences in hydrolytic activities between the variants were likely caused by an unpredicted and complicated folding of each lipase protein [21]. The mutation at the oxyanion Q114 could have simply modified the topology and the binding capacity within the locality of the active site that

Figure 2. The effect of the various natural oils on the activity of the lipase variants, Phe. Met, Ile, Leu, Val, Trp and wildtype T1, arranged in descending order of hydrophobicity of the substituting residues. The reaction mixtures were assayed for 30 min with agitation at 200 rpm. Each mixture was prepared in triplicates, in two determinants and the results were the mean of the obtained experimental reading.
influenced the distances from the nucleophile to the acid/ base catalyst and to the substrate [22].
3.4. Effect of Organic Solvents
Figure 3 compares the effect of the solvent (log P) on the activity of the variants and wild-type. In DMSO (log P –1.38) and acetonitrile (log P –0.40), the Leu variant exhibited higher activity than the wild-type, although lipase activity was marginally higher in acetonitrile. Substituting Leu, Met and Phe into Q114 increased enzyme activity in ethyl acetate but activity of the Trp variant was adversely affected. Interestingly, the variants largely exhibited significantly lowered activity when supplemented with solvents of moderate log P, ranging from 2.50 to 4.40. The solvents which had negative impact on the variants were toluene (log P 2.50), hexane (3.50) and isooctane (log P 4.40), respectively.
However, enzyme activity generally improved in solvents with higher log P, namely, in N-tetradecane (log P 7.60) and N-hexadecane (log P 8.80), especially in the latter. Four of the lipase variants performed better than the wild-type in N-hexadecane, except for lipase variants Val and Trp. Remarkably, substitution of Leu improved the activity of the enzyme over the wild-type in five out of the eight tested solvents. This indicated that Leu affected the lipase activity in these solvents differently from the wild-type. As earlier observed in solvents with lower log P, substitution with Trp and Val had significantly increased susceptibility of both variants to solvent inhibition.
The log P is a term that is used to demonstrate the solvent-effect on the activity of enzymes. It is defined as the logarithm of the partition coefficient in a standard noctanol/water two phase system [23]. The increased activity of the Leu variant towards the water-miscible solvents, DMSO, acetonitrile and ethyl acetate could perhaps be associated to an increased hydrophobic interaction at the catalytic site [24], brought about by the hydrophobic Leu residue. As the log P of solvents was increased, the activity of the lipase variants were largely improved. In the presence of higher log P solvents, the enzyme molecules were less susceptible to stripping of essential water molecules when exposed to hydrophobic solvents versus hydrophilic solvents [25]. Therefore, the native form of the active site was maintained and the enzyme kept its catalytically competent form. Therefore, high catalytic efficiency in the lipase variants could be achieved.
According to previous reports, solvents with log P values in the range of 1.5 to 4.0 are severely toxic because they preferentially partitioned in the cytoplasmic membrane [26]. The findings in this study concurred with previous reports on similar lipase inhibitions caused by solvents within this range. The log P values of solvents, toluene, hexane and isooctane were well within this range which probably the reason behind the considerable decline in the activity of the lipase variants. These solvents could interfere with the intricate balance between the hydrophobic interactions, hydrogen bonds and van der Waals interactions that were essential for proper protein folding and native protein structure [27].
3.5. Effect of Surfactants
The effect of the various surfactants on the activity of the lipase variants and wild-type is shown in Figure 4. Substitution of Q114 with Leu considerably improved its activity over the wild-type in four different surfactants, namely, Tween 40, Tween 80, Span 20 and SDS, respectively. The highest activity was recorded for the Leu variant in Span 20. Replacement with Val slightly enhanced its activity in Span 20 but also increased susceptibility to inhibition by Tween 20 - 60. Lipase activity
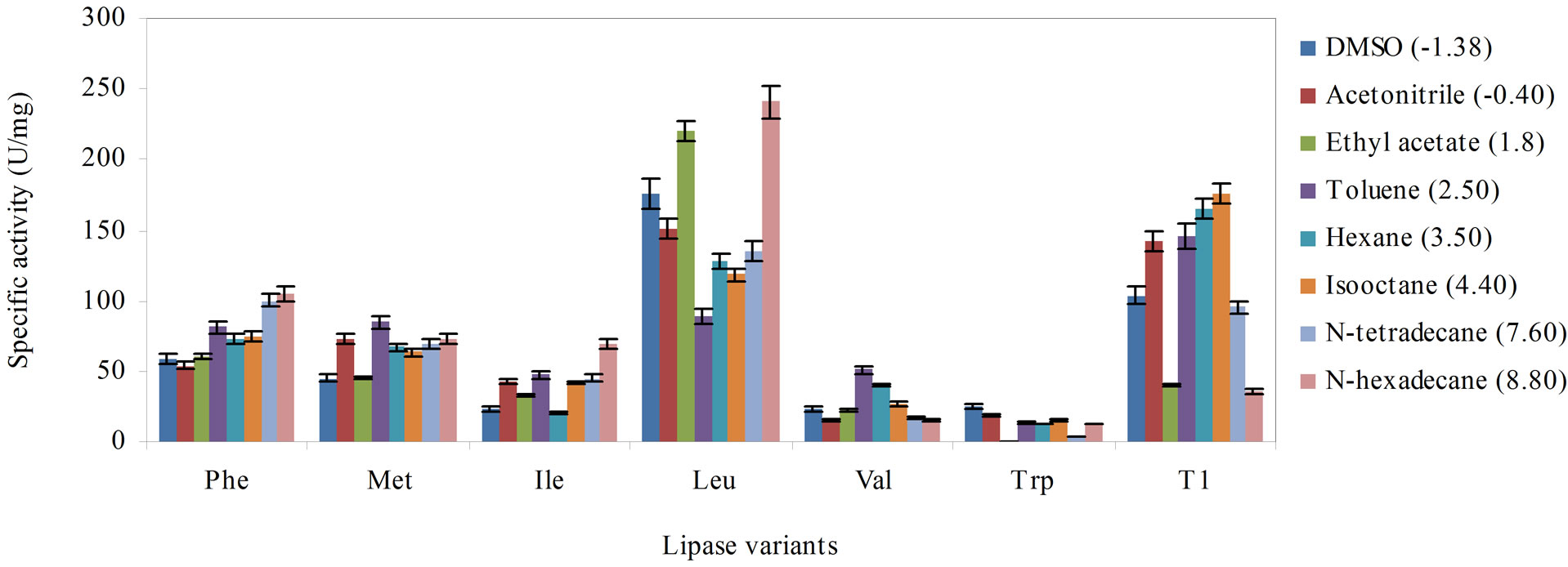
Figure 3. The effect of the various solvent hydrophobicity on the stability of the lipase variants, Phe, Met, Ile, Leu, Val, Trp and wild-type T1 lipase to catalyze hydrolysis of olive oil, arranged in descending order of hydrophobicity of the substituting residues. The lipase variants were treated with the solvents (1:3 v/v) for 30 min at 30˚C and were subsequently assayed 30 min with agitation at 200 rpm. Each mixture was prepared in triplicates, in two determinants and the results were the mean of the obtained experimental reading.

Figure 4. The effect of the various surfactants on the stability of the lipase variants, Phe, Met, Ile, Leu, Val, Trp and wildtype T1 lipase to catalyze hydrolysis of olive oil, arranged in descending order of hydrophobicity of the substituting residues. The lipase variants were treated with the surfactants (0.1% v/v) and incubated for 30 min with agitation at 200 rpm. Each mixture was prepared in triplicates, in two determinants and the results were the mean of the obtained experimental reading.
was significantly improved in SDS for the Met, Ile, and Leu variants. On the other hand, substitution with Trp and Ile resulted in both variants to be easily inhibited by the various surfactants. The specific activity in the Trp and Ile variants were less than 30% of the wild-type, except the latter was more stable in SDS.
Surfactants are amphipathic molecules with a hydrophilic head group and a hydrophobic tail [28]. The diminished catalytic activity of the Leu variant in Tween 20 and the Val variant in SDS, followed a similar trend of deactivation as previously reported for BTL2 lipase from Geobacillus thermocanulatus. The BTL2 lipase was totally inhibited by the same concentration of surfactants, Tween 20 and SDS, respectively [29]. Meanwhile, the higher lipase activity in SDS for the variants, Ile, Met and Leu implied that SDS might have increased adsorption of the lipase molecules to the water-lipid interface. This could have facilitated contact between the enzyme and substrates that enhanced catalysis. Additionally, SDS could have also invoked changes in lipase conformation [30] and caused the enzyme structure to revert to a more catalytically active form.
When compared to Tween 20 - 80 group, SDS was considerably more disruptive on the structure of the lipases. The inhibitory effects of SDS could be attributed to the ionic surfactant binding to the hydrophobic and hydrophilic heads in the affected variants. This caused the varied degrees of unfolding of their tertiary structures and lowered their catalytic activity [31]. This phenomenon, however, was not seen in Tween 20 - 80 and Span 20 as

Figure 5. The effect of the various metal ions on the activity of the lipase variants, Phe, Met, Ile, Leu, Val, Trp and wildtype T1 lipase to catalyze hydrolysis of olive oil, arranged in descending order of hydrophobicity of the substituting residues. The lipase variants were treated with the metal ions (1 mM) and incubated for 30 min with agitation at 200 rpm. Each mixture was prepared in triplicates, in two determinants and the results were the mean of the obtained experimental reading.
their rigid and bulky non-polar heads could not enter into the water-soluble protein [27,32]. Meanwhile, Tween 20 - 80 which inhibited the lipase variants implied these surfactants might have triggered the opening of the lid present on the lipases, either by water-substrate interface or detergent-micelles. Therefore, monomers of non-ionic detergents could still enter and their molecules bound to the active site that caused the enzyme inhibition [33].
3.6. Effect of Metal Ions
Figure 5 depicts the effect of various metal ions on the activity of the lipase variants and wild-type. The activity of the lipase variants were generally promoted by divalent metals, namely, Sr(II), Ca(II) and Mn(II). Interestingly, we found Sr(II) enhanced hydrolysis in variants Val, Ile, Met and Phe, as well as the wild-type. The highest activity was noted in the Leu variant in reactions supplemented with Sr(II), followed by reactions containing Ca(II). Comparable lipase activity as the wild-type was observed in the Val substituted variant, however, activity was considerably lower in mixtures containing Ca(II). On the other hand, reactions supplemented with Li(I) and K(I) ions showed significant loss of lipase activity in all variants. Supplementation with the various metal ions failed to promote catalysis in the Trp variant. In general, the Trp variant exhibited the lowest specific activity in almost all of tested metal ions.
Metal ions can act as electrophiles that seek the possibility of sharing electron pairs with other atoms, so that a bond or a charge-charge interaction could be formed [34]. Ca(II) ions was reported to confer stabilizing effect on the wild-type lipase [8]. However, activity of the lipase variants were largely improved in media supplemented Sr(II) ions suggesting that Sr(II) could be used to promote reactions catalyzed by these lipases. Sr(II) ions probably stabilized the lipase protein structures by retaining the catalytic residue at the correction position during elevated temperatures. This prevented the lipase variants from undergoing conformational changes during catalysis. Conversely, the notably reduced lipase activity in Li(I) and K(I) supplemented reactions could be the result of undesirable interactions between the side chains of the surface amino acids and the charged salt ions [26,35]. This might have negatively influenced the protein conformational stability of the variants. The interactions between the salt ions and enzyme surfaces were proposed to affect ionization of certain amino acid residues and alter enzyme activity altogether [36].
4. CONCLUSION
Our investigation has successfully elucidated the role of the oxyanion 114 was not exclusive to just the binding with the substrate during catalysis but also contributed to conformational stability and conferring specific catalytic attributes to T1 lipase. Substitution of Leu into Q114 afforded the largest improvement in the catalytic attributes. Significant improvements in conformational stability and catalytic attributes were observed for the Leu variant in four out of the six tested characteristics. Replacement with Met resulted in a lipase variant that was more efficient in hydrolyzing a wide range of natural oils than the wild-type. These results exemplified that a single point residue changes at the oxyanion of an enzyme could create improvements in a myriad of enzyme attributes, which implied there was some interplay between hydrophobicity and conformation for lipase catalytic functions.
5. ACKNOWLEDGEMENTS
This work was supported by a grant of the Ministry of Science, Innovation and Technology of Malaysia, Vot number 5487729.
NOTES