Composition and Diversity of Soil Bacterial Communities along an Environmental Gradient in the Sudano-Sahelian Region of Senegal ()
1. Introduction
Soil microorganisms represent most biodiversity in terrestrial ecosystems [1]. As the most abundant groups of soil microbes and primary consumers in the soil food web [2], soil microorganisms such as bacteria and fungi play critical roles in regulating soil fertility, plant health, and soil nutrient cycling [3]. Despite their key role in soil biofunctioning that ensures many soil ecosystem services, studies on soil biodiversity are often overlooked [4], more so in drylands than in other ecosystems [5].
In semi-arid region, the Sudano-Sahelian zone (SSZ) of West Africa consists of two roughly parallel ecological regions, the Sahel and the Sudan [6]. The Sahel is located between the humid equatorial zone of Africa to the south and the Sahara desert to the north, within the semiarid to arid transition zone [7]. As one of the world’s largest dryland areas [8], the Sahelian region has a very strong north-south gradient with between 200 mm and 600 mm annual rainfall [9] while the Sudanian zone receives between 600 and 1000 mm per year [10]. With its strong east-west uniformity of climate and vegetation conditions [9], the Sudano-Sahelian zone of West Africa (SSZ) presents diverse land covers. The northernmost areas of the Sahelian region are characterized by sparse vegetation and grassland interspersed with cropland while in the Sudanian zone in the south, shrublands, dense woody savannas and forest mosaics predominate [8] [11] [12]. The region is characterized by high rainfall variability [13] - [18] and has experimented severe and longest droughts during the twentieth century [19] [20] with persistent dry conditions since then [19]. In a context of climatic fluctuations and anthropogenic disturbances leading to land degradation in the region and where the impacts are most apparent in the surface vegetation cover [21], studies have mainly focused on the dynamics of vegetation and land cover changes [22] - [28]. While the relationship between vegetation and rainfall in these semi-arid environments has been extensively studied [5] [29], soil microbial communities remain largely unexplored.
Limited soil water availability (SWA) can impair the function of soil ecosystems long before symptoms become visible aboveground [30]. It is well known that a shift in the rainfall regime can directly affect soil microbial communities by altering soil water content, and indirectly by changing other physico-chemical features [31]. At regional scale, climate and soil parameters are usually identified as key predictors of microbial community structure [32]. Community structure and spatial variations in soil microbial communities have been documented at large spatial scales in different ecosystems and biomes ranging from polar [33] [34] to desert [35] [36] and including grasslands [37], steppes [38], forests [39], savannah [40]. Some soil properties and environmental variables such as precipitation [41], vegetation [42] [43], land use [44], pH [45], soil texture [46], carbon and nutrient content [47], soil moisture [48] have been described as potential driving factors of shifts in microbial communities [49].
An understanding of the factors that influence the biodiversity of soil bacterial communities is needed, first as a framework for determining the roles of different taxa in soils; and also to predict ecosystem responses to a changing environment [50]. In this work the objective was to explore in Senegal, a country partly straddling the Sudano-Sahelian zone (SSZ) of West Africa: 1) the composition and diversity of the soil bacterial communities and 2) to identify and prioritize among environmental parameters potential driving factors of the composition of bacterial communities in this semi-arid environment. Non-anthropogenic sites across four pedoclimatic zones forming an environmental gradient were sampled. We hypothesized that diversity and composition of the soil bacterial communities will vary along the environmental gradient, in response to climate and soil characteristics.
2. Material and Methods
2.1. The Study Area Description
The study was conducted in the Sahelian, Sahelo-sudanian and Sudanian climatic zones of Senegal (12˚30' - 16˚30'N; 11˚30' - 17˚30'W) a country located in the extreme western part of the African continent (Figure 1). Senegal has a semi-arid tropical climate with a cold dry season lasting from November to April and a hot rainy season from May to October. Soils range from dry sandy soils in the north to tropical ferruginous soils in the central region and to ferralitic highly leached soils in the south [51]. Rainfall distribution is characterized by an increase in the amount of precipitation and number of rainy days from the north to the south [52], resulting in a north–south spatial precipitation gradient. This pronounced climate gradient has led to the establishment of different climatic zones geographically defined on the basis of long-term precipitation averages, with a latitudinal pattern of vegetation types and ecosystems. Referring to [53] paper that provided ecological stratification of the Senegalese country in different ecoregions with overview of historical land use and land cover trends:
- The Sahelian climatic zone is a hot and dry semi-arid region that covers the northern part of the country. The climate is continental Sahelian with average annual rainfall below 400 mm. The area includes the Senegal River Valley and a large sylvo-pastoral sandy area known as sandy “Ferlo” with ferruginous tropical soils and representing the Northern pastoral ecoregion. Vegetation is generally characterized by open herbaceous types (steppe and short grass savannah) often mixed with woody plants.
- The Sahelo-Sudanian climatic zone has acontinental Sahelian to Sahelo-Sudanian climate with average annual rainfall between 400 and 600mm. This climatic area includes mainly a West Central Agricultural Region with ferruginous tropical sandy soils slightly leached and pastoral areas formed at the West by the Southern Pastoral Region with sandy soils (Sandy “Ferlo”) and at the East by the Ferruginous pastoral ecoregion (or Lateritic “Ferlo”) with shallow, loamy, gravelly soils. Vegetation is characterized by shrubby savannah.
- The Sudanian climatic zone located in the southern half of the country present a coastal Sudanian climate in the west and continental Sudanian climate in the center and east. Average annual rainfall ranges from 600 to 800 mm. The rainfall gradient is not north-south, which is the case at the two other sites but rather east to west. The Sudanian zone present two agricultural ecoregions at the West and an Eastern Transition Region with shallow loamy and gravelly soils over laterite on plateaus; sandy to loamy leached ferruginous soils in the valleys and terraces. The natural vegetation varied from degraded shrub and tree savannas to wooded savannah.
Details on the different ecoregions of the different climatic zones can be found in [53] paper.
2.2. Soil Sampling Strategy
Four sampling zones a priori different in climate and soil conditions were considered across the Sahelian, the Sahelo-sudanian and the Sudanian climatic zones (Figure 1). The first sampling area was located at the northern–west part of the Sahelian climatic zone. The second and the third sampling areas were located respectively at the northwestern part and the northeastern part of the Sahelo-sudanian climatic zone. The last and fourth sampling zone concerned the eastern part of the Sudanian climatic zone.
![]()
Figure 1. Map of the study area showing the location of the sampling sites across the sampling zone. Sahelian zone transect (●), Western Sahelo-Sudanian zone transect (×), Eastern Sahelo-Sudanian zone transect (■) and Sudanian zone transect (♦). Average annual precipitation from the WorldClim 2 data collection [54].
Sampling was conducted at the end of the rainy season in November 2015. Using transect sampling strategy, soil samples were collected from random sampling sites along each sampling transect (national roads) with a total of 24 sampling sites. Samples were taken under similar vegetation cover (natural herbaceous cover) where no livestock grazed and no known human activities took place. In each sampling site or plot, one composite sample was collected from 5 core samples at 0 - 10 cm soil depth and 1 m apart, using a cylindrical soil sampler (5 cm inner diameter, 10 cm length). Composite samples were immediately placed in plastic bags and stored at 4˚C in a cooler. Other soil cores were also collected for the determination of bulk density. Back to the laboratory coarse plant debris were removed prior to a gently air-drying and sieving at 2 mm. Subsamples for molecular analyses were stored at −80˚C while subsamples for the determination of the physico-chemical analyses were air-dried and stored. Ecological characteristics and details of the sampling zones are summarized in Supplementary TableS1.
2.3. Extraction of the Climate Data
Mean Annual Temperature (MAT) and Mean Annual Precipitation (MAP) of each sampling site was extracted based on geographic coordinates (Latitude & Longitude). Values extracted represented averages for the period (1970-2000) of bioclimatic variables of WorldClim 2 data collection [54] using BIO1 (Annual Mean Temperature) and BIO12 (Annual Precipitation) variables at a resolution of 1 km (~30 arc-seconds).
2.4. Soil Physical and Chemical Analyses
Soil texture was determined by sedimentation using Robinson’s technique [55]. Soil moisture content was calculated as follows: (mass of wet soil – mass of dry soil)/(mass of dry soil). Dry mass was determined from the mass of wet soil after drying at 105˚C/for 48 h in an oven. Bulk density (g∙cm−3) was determined by the ratio (dry mass of volumetric sample)/(volume of the cylindrical soil sampler). Soil pH was quantified in a water soil suspension [soil: water ratio 1:2.5 (w/v)] (InoLab 720, WTW, Germany). Soil inorganic N (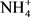 -N and
-N and 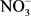 -N) was extracted from 10 g of fresh soil with 2 M KCl solution and measured using a colorimetric method with a flow injection auto-analyzer (SEAL analytical, Auto Analyzer 3-QuAAtro, Seal Analytical, France). Soil total nitrogen and carbon were quantified by combustion using ThermoFinnigan Flash EA 1112 (ThermoFinnigan, France). All these soils analyses were conducted at the Laboratoire des Moyens Analytiques (LAMA), a laboratory belonging to the Institut de Recherche pour le Développement (IRD) in Dakar, Senegal.
-N) was extracted from 10 g of fresh soil with 2 M KCl solution and measured using a colorimetric method with a flow injection auto-analyzer (SEAL analytical, Auto Analyzer 3-QuAAtro, Seal Analytical, France). Soil total nitrogen and carbon were quantified by combustion using ThermoFinnigan Flash EA 1112 (ThermoFinnigan, France). All these soils analyses were conducted at the Laboratoire des Moyens Analytiques (LAMA), a laboratory belonging to the Institut de Recherche pour le Développement (IRD) in Dakar, Senegal.
2.5. DNA Extraction and 16S rRNA Gene Sequencing
Total genomic DNA of each soil sample was extracted from 0.25 g of soil using the FastDNA™ SPIN kit for Soil (MP Biomedicals, CA, USA) in accordance with the manufacturer’s instructions. DNA extracts were purified by adding 500 μl of guanidine thiocyanate (5.5M) and then centrifuged at 14000 g for 5 min at 4˚C. DNA was suspended in 150 μL elution buffer and quantified with a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA) before being stored at −20˚C in the freezer. Sequencing of gene encoding 16S rRNA was performed at MRDNA (http://www.mrdnalab.com/, Shallowater, TX, USA) with an Ion Torrent PGM system for sequencing (Life Technologies Corp., Thermo Fischer Scientific, Massachusetts, USA). Briefly, samples were barcoded and the V4 variable region of 16S rRNA gene was targeted with the 515F/806R primer set [56]. PCR amplification was carried out using the HotStar Taq plus Master Mix Kit (Qiagen, Germantown, MD, USA) under the following conditions: one cycle at 94˚C for 3 min, followed by 28 cycles at 94˚C for 30 s, at 53˚C for 40 s and at 72˚C for 1 min plus a final elongation step at 72˚C for 5 min.
2.6. Sequence Analysis Processing and OTU Clustering
Barcodes and primers were removed from the sequences. Short sequences < 150 bp, sequences with ambiguous base calls and sequences with homopolymer runs exceeding 6 base pairs were removed. The remaining sequences were then denoised and operational taxonomic units (OTU) were defined by clustering at 3% divergence (97% similarity) followed by removal of singletons and chimeras. Final OTUs were taxonomically classified using BLASTn against a curated database derived from GreenGenes, RDPII and NCBI (http://www.ncbi.nlm.nih.gov/, [57], http://rdp.cme.msu.edu). Only OTUs identified up to at least the phylum level were kept on the final OTUs tables. The final OTUs tables containing the number of sequences per sample per OTU matching the designated taxonomic classification were then produced. The whole process was conducted at MR DNA using the MR DNA ribosomal and functional gene analysis pipeline (http://www.mrdnalab.com/, MR DNA, Shallowater, Texas, USA). The raw sequence reads were submitted to the NCBI SRA database under accession number PRJNA508511.
2.7. Statistical Analysis
2.7.1. Analysis of Bacterial Diversity
The analyses of the OTU tables were performed in R [58] using R phyloseq package [59]. OTU tables were first randomly subsampled according to the number of sequences obtained from the smallest library in order to deal with unequal sampling effort. We arbitrarily defined “Rare” and “Abundant” OTUs as follows: OTUs whose reads summed across all samples was less than 0.01% of the total number of bacterial sequences were defined as “rare taxa” while OTUS with total relative abundance greater than 0.01% were considered “abundant”. Taxonomic composition and taxa relative abundances of the bacterial communities were determined by aggregating sequence counts of OTUs sharing the same taxon. Bacterial α-diversity was evaluated for each soil sample by calculating richness and the Shannon and Simpson indexes using R Vegan software package [61]. One way analysis of variance (ANOVA) was used to compare α-diversity between the pedoclimatic zones with XLSTAT software [60]. Changes in bacterial community composition (β-diversity) between soil samples were assessed using a non-metric multidimensional scaling analysis (NMDS) based on the Bray-Curtis distance index calculated at the OTU level. NMDS ordination was then tested by PERMANOVA (distance-based permutational multivariable analysis of variance) using 9999 permutations with the Adonis function in the R Vegan package [61].
To identify the features (OTUs) that drive the structure of the bacterial community, sparse partial least square discriminant analysis (sPLS-DA) of abundant OTUs was conducted using the R mixOmics package [62]. sPLS-DA aims to identify a small subset of genes that best discriminate the groups according to a given factor by including a LASSO penalization to select the most informative predictors. Partial least squares discriminant analysis (PLS-DA) is an exploratory approach that seeks the optimal linear combinations of variables (genes) that best separate the sample groups [63]. The classification performance (classification error rate) of the model for the sPLS-DA were evaluated using the R perf function with a leave-one-out cross validation. Final predictors (OTUs) were grouped at the genus level to display taxonomic labeling.
2.7.2. Relationship between Environmental Parameters
Climate data and soil physical-chemical properties of the sampling zones were compared using Welch-ANOVA test with a significance level of 5%. Games-Howell test were carried out to test for significant differences between treatments. The differences were considered significant at P < 0.05. Correlations between environmental variables were assessed by Pearson’s test. All the statistical analyses were carried out using R XLSTAT software [60].
2.7.3. Relationship between Bacterial Diversity and Environment Variables
The relationship between bacterial diversity and environment variables was assessed by performing a distance-based redundancy analysis (db-RDA). The bacterial community composition matrix was generated from the OTU table using the Bray-Curtis dissimilarity index and the environmental data were first standardized by centering and scaling to avoid the effects of scale between the explanatory variables. The relative contribution of each significant explanatory variable to the fraction of variance explained by the environmental data was determined by analysis of variance (ANOVA)-like permutation test for canonical analyses (anova.cca function in the vegan package). All statistical analyses were performed on the R statistical platform using the vegan package [61].
3. Results
3.1. Environmental Parameters
Mean annual temperature (MAT) and Mean Annual Precipitation (MAP) varied significantly between the soil sampling zones (Table 1). MAT ranged from 25˚C to 28˚C while MAP ranged from 290.7 to 638.8 mm. Soil textural classification revealed that the north-west Sahelian zone and the western Sahelo-Sudanian zone were characterized by sandy soils with more than 90% of sand. The eastern Sahelo-Sudanian and the Sudanian zones were more clayey (over 7%) and silted (over 15%) and comprised respectively loamy sand and sandy-loam soils (Table 1). As expected climate and soil texture characteristics differentiated our four sampling zones a priori different in soil and climate conditions in four pedoclimatic zones: a Sahelian sandy zone (SS), a Sahelo-Sudanian sandy zone (SSS), a Sahelo-Sudanian loamy-sand zone (SSLS) and a Sudanian sandy-loam zone (SSL).
Physico-chemical parameters varied in space with significant differences among the pedoclimatic zones (Table 2). Mean soil pH varied significantly between the soil samples and ranged from 5.72 to 7.43 across the sites. The SS soil samples were characterized by neutral pH while the others soil samples were slightly more acidic. Soil moisture, total C, total N and mineral N contents were
![]()
Table 1. Climatic and textural characteristics of the sampling zones.
Values are means (Standard deviation). Values with different letter are significantly different (P < 0.05).
![]()
Table 2. Soil physicochemical properties across the pedoclimatic zones.
Values are means (Standard deviation). Values with different letter are significantly different (P < 0.05). N_min = Mineral nitrogen, C/N = carbon/nitrogen ratio, CEC = Cation exchange capacity.
significantly higher in the SSL soil samples than in the SS, SSS and SSLS samples (Table 2). Only soil bulk density, CEC and the C/N ratio did not change significantly across the pedoclimatic zones. Pearson’s correlation coefficient (Figure 2) revealed that MAT and MAP were both significantly correlated with clay and silt contents, total C, total N, mineral N, CEC and soil moisture, which in turn, also showed a positive correlation among them. In contrast, those soil properties were significantly negatively correlated with soil pH, which in turn, was highly correlated with soil sand contents. Our results showed the study area to be clearly marked by a soil texture gradient consisting of an increase in clay, silt, total C, total N, mineral N, CEC and soil moisture contents and a decrease in pH and sand content from the Sahelian to the Sahelo-Sudanian zone and to the Sudanian zone.
3.2. Taxonomic Composition of Soil Bacterial Communities
Across the 24 soil samples, a total of 619,200 high-quality sequences were obtained and clustered in 17,253 bacterial OTUs. The majority of the OTUs i.e. 89% were present in low abundance while the abundant OTUs represented only 11% of the OTUs (Supplementary TableS2).
The taxonomic inventory of the bacterial sequences identified 24 phyla. The dominant bacterial phyla (Figure 3) were Firmicutes (30.42%), Proteobacteria
![]()
Figure 2. Pearson’s correlations coefficients between the soil environmental (climatic and edaphic) variables. Positive significant relations are in blue and negative significant relations are in red, with the intensity of the relation changing from dark to light.
![]()
Figure 3. Taxonomic composition and relative abundance of the soil bacterial communities at the phylum level. Sites 1 to 4 = Sahelian sandy zone; Sites 5 to 13 = Sahelo-sudanian sandy zone; Sites 14 to 18 = Sahelo-sudanian loamy sand zone; Sites 19 to 25 = Sudanian sandy loam zone. Only top (relative bacterial abundance > 1%) bacterial taxa are shown.
(24.33%), Actinobacteria (23.44%), Chloroflexi (6.77%), Gemmatimonadetes (4.46%), Acidobacteria (3.81%), Planctomycetes (2.88%), Verrucomicrobia (1.5%), Bacteroidetes (1.34%). These nine dominant phyla accounted for more than 98% of all the bacterial sequences. Rare phyla (relative bacterial abundance < 1%) were represented by Cyanobacteria, Nitrospirae, Armatimonadetes, Deinococcus_thermus, Chlamydiae, Fibrobacteres, Fusobacteria, Ignavibacteriae, Synergistetes, Tenericutes, Elusimicrobia, Nitrospinae, Chrysiogenetes, Chlorobi and Thermodesulfobacteria.
At the class level, 50 bacterial classes belonging mainly to Firmicutes, Proteobacteria and Actinobacteria phyla were found across the samples. The most frequent were Bacilli (27.3%), Actinobacteria (22.4%), α-Proteobacteria (13.1%), d-Proteobacteria (6.1%), Gemmatimonadetes (4.5%), b-Proteobacteria (3.5%), Ktedonobacteria (3.2%), Clostridia (3.1%), Planctomycetia (2.6%), Acidobacteriia (2.1%), γ-Proteobacteria (1.7%), Chloroflexia (1.6%), Verrucomicrobiae (1.4%) and Thermoleophilia (1%) classes (Figure 4).
At low taxonomic level (genus level), 251 genera were detected. The most important (relative sequence abundance greater than 1%) were Bacillus (17.4%), Conexibacter (4.9%), Ammoniphilus (3.4), Solirubrobacter (3.1%), Microvirga (1.8%), Rubrobacter (1.7%), Streptomyces (1.6%), Paenibacillus (1.5%), Pelobacter (1.4%), Sphingomonas (1.3%), Bradyrhizobium (1.2%), Gemmatimonas (1.1%), Acidobacterium (1.1%), Kaistobacter (1.0%) (Figure 5). Proportion of bacterial sequences unidentified at genus level represented 24.5% of the total bacterial sequences.
Comparison of the composition of the bacterial communities at the different
![]()
Figure 4. Taxonomic composition and relative abundance of the soil bacterial communities at the class level. Sites 1 to 4 = Sahelian sandy zone; Sites 5 to 13 = Sahelo-sudanian sandy zone; Sites 14 to 18 = Sahelo-sudanian loamy sand zone; Sites 19 to 25 = Sudanian sandy loam zone. Only top (relative bacterial abundance > 1%) bacterial taxa are shown.
![]()
Figure 5. Taxonomic composition and relative abundance of the soil bacterial communities at the genus level. Sites 1 to 4 = Sahelian sandy zone; Sites 5 to 13 = Sahelo-sudanian sandy zone; Sites 14 to 18 = Sahelo-sudanian loamy sand zone; Sites 19 to 25 = Sudanian sandy loam zone. Only top (relative bacterial abundance > 1%) bacterial taxa are shown.
taxonomic level across the different pedoclimatic zones showed that the composition at low taxonomic level (genus level) was more spatially structured with clear distribution patterns of the relative abundance of some bacterial taxa, than at higher taxonomic level (phylum and class).
3.3. Bacterial α and β-Diversity
Our results showed that bacterial α-diversity was stable across the different pedoclimatic zones (Table 3). OTU richness was similar between the different pedoclimatic zones. The specific diversity based on the values of the Shannon and Simpson diversity indices, was high with codominance of species but like OTU richness did not vary significantly between the different pedoclimatic zones.
Changes in the bacterial community composition among the different groups of soil samples were assessed using NMDS ordination at the OTU level. Non-metric multi-dimensional scaling clustered the soil samples according to their pedoclimatic origin (Figure 6(a)). NMDS analyses without the rare OTUs did not alter the structuring patterns of the bacterial β-diversity (Figure 6(b)). Dissimilarities between soils groups seemed to be conserved across the two ordinations results (NMDS with and without the rare fraction). NMDS clustering was statistically supported by the PERMANOVA analysis (adonis R2 = 0.27, p < 0.001) and confirmed that significant dissimilarities occurred in the composition of the bacterial communities.
3.4. Predictors of the Structuring Patterns of Bacterial β-Diversity
To identify OTUs involved in bacterial β-diversity structuring patterns, a
![]()
Table 3. Bacterial community richness and diversity indices across the pedoclimatic zones.
Values are means (Standard deviation). Values with different letter are significantly different (P < 0.05).
![]() (a) (b)
(a) (b)
Figure 6. NMDS ordination of the soils bacterial communities. (a) The rare fraction community was included in the analysis. (b) The rare fraction community was excluded from the analysis. Each point represents a soil sample community: Sahelian sandy zone (Red circles); Sahelo-sudanian sandy zone (Green circles); Sahelo-sudanian loamy sand zone (Green circles), Sudanian sandy loam zone (Purple circles).
sPLS-DA analysis was performed on the abundant fraction of the bacterial community at the OTU level. The sPLS-DA analysis distinguished the soil samples according to their pedoclimatic origin (Figure 7(a), Figure 7(b)), confirming the significant changes in the bacterial community composition between the soil groups. The four (4) pedoclimatic zones defined in the study were clearly separated by the first three PLS components with respectively 23%, 12% and 6% of the variance. Numerous unique OTUs (1,311 OTUs) were identified as drivers of the clustering of the soil samples across the three components of the sPLS-DA.
The first sPLS-DA component with a selection of 429 OTUs mainly separated the Sahelian sandy zone (SS) from soils of other areas and more particularly those from the Sudanian sandy-loam zone (SSL). OTUs characterizing the Sahelian sandy zone (SS) were predominantly assigned to the genus Bacillus, Rubrobacter, Microvirga, Pelobacter, Ammoniphilus, while Solirubrobacter, Conexibacter, Kaistobacter, Bacillus and Sphingomonas were mainly associated with the Sudanian sandy-loam zone (SSL).
The second sPLS-DA component with 419 discriminating OTUs separated the soils of the sandy Sahelo-Sudanian zone (SSS) but also those of the Sudanian sandy-loam zone (SSL) from the soils of the others pedoclimatic zones. The OTUs characterizing the sandy Sahelo-Sudanian zone (SSS) were mainly affiliated to the genera Conexibacter and Bacillus while those (160 OTUs) characterizing the Sudanian sandy-loam zone (SSL) belonged to the genera Kaistobacter, Sphingomonas, Bacillus and Conexibacter. The third and last sPLS-DA component, with a selection of 703 OTUs, separated the soils of the Sahelo-Sudanian loamy-sand zone (SSLS) from the others soil groups. The OTUs characterizing this pedoclimatic zone belonged mainly to the genus Conexibacter and Solirubrobacter. All the predictors OTUs characterizing each pedoclimatic zone were displayed on a heatmap (Figure 7(c)) highlighting changes that occurred in the composition of the soil bacterial communities across the four pedoclimatic zones.
The results of the sPLS-DA analysis clearly showed that the structuring patterns of the bacterial β-diversity across the pedoclimatic zones were driven by shifts in the relative abundance of OTUs belonging mainly to the genera Bacillus, Rubrobacter, Microvirga, Pelobacter, Solirubrobacter, Conexibacter, Kaistobacter and Sphingomonas. Assessment of the spatial distribution of the abundance of the said genera across the pedoclimatic zones confirmed the distribution patterns previously mentioned. Conexibacter and Solirubrobacter genus belonging to the Actinobacteria class showed increasing abundances from the dry Sahelian sandy zone (SS) to the more humid Sudanian sandy-loam zone (SSL) whereas the abundance of Rubrobacter genus belonging also to Actinobacteria decreased (Supplementary FigureS1). Similarly, Kaistobacter and Sphingomonas belonging to the α-proteobacteria class showed increasing abundances from the dry Sahelian sandy zone (SS) to the more humid Sudanian sandy-loam zone (SSL) while Microvirga also affiliated to α-proteobacteria showed the
![]() (a) (b)
(a) (b)![]() (c)
(c)
Figure 7. sPLS-DA score plot distinguishing soil bacterial communities according to the pedoclimatic zone on the first (a) and second component (b) of the sPLS-DA analysis. (c) Clustered Image Map (CIM) of the OTUs selected on the three components of the sPLS-DA: the heatmap highlights the OTUs that best characterize the different soils groups (pedoclimatic zones). Soil samples clusters are indicated on the left-hand side of the heatmap and OTUs clusters on the top.
opposite trend (Supplementary FigureS2). Pelobacter genus affiliated to δ-proteobacteria class rather showed decreasing abundance from the dry Sahelian sandy zone to the Sudanian sandy-loam zone (SSL) (Supplementary FigureS3).
Finally, genus Bacillus did not show spatial variation; the genus predominated in all soils regardless of their pedoclimatic origin (Supplementary FigureS5).
Additionally, these results indicated that these genera driving the structuring patterns of the bacterial β-diversity along the pedoclimatic gradient belonged mainly to Bacilli, α-Proteobacteria, d-Proteobacteria, Ktedonobacteria and Actinobacteria classes with some bacterial genera belonging to the same phylum but with divergent distribution patterns of their relative abundance.
3.5. Relations between Bacterial Community Composition and Environmental Variables along the Climatic Gradient
Distance-based redundancy analysis (db-RDA) was used to determine the influence of environmental variables on bacterial community composition along the pedoclimatic gradient. Soil pH, moisture and clay content were the only environmental parameters that were significantly correlated with the bacterial β–diversity patterns (Supplementary TableS3). These three variables explained 25.5% of the dissimilarities between the bacterial communities with the soil pH as the main driver, followed by soil moisture and then by soil clay content.
The db-RDA plot (Figure 8) displayed these significant relations. The first db-RDA axis related to soil pH, showed clearly a structured pattern of the bacterial communities. Bacterial communities from the Sahelian sandy soils, characterized by neutral pH, were separated from bacterial communities of the others pedoclimatic zones characterized by more acidic pH values. The second db-RDA axis correlated with soil moisture and soil clay contents formed two clusters separating the bacterial communities of the sandy Sahelian and Sahelo-Sudanian zones from those of the Sahelo-Sudanian loamy-sand zone (SSLS) and Sudanian sandy-loam zone (SSL).
4. Discussion
4.1. Taxonomic Composition of the Soil Bacterial Communities
Taxonomic composition of soil bacterial communities of the Sudano-sahelian region of Senegal revealed the predominance of Firmicutes, Chloroflexi and Gemmatimonadetes besides Acidobacteria, Proteobacteria and Actinobacteria phyla generally considered as the most abundant in soils [45] [64]. This finding agrees with those of the rare studies on microbial communities conducted in
![]()
Figure 8. Distance based redundancy analysis (db-RDA) showing relations between bacterial community structure and explanatory variables. Each point represents a soil sample community: Sahelian sandy zone (Yellow circles); Sahelo-sudanian sandy zone (Orange circles); Sahelo-sudanian loamy sand zone (Brown circles), Sudanian sandy loam zone (Green circles).
tropical Africa. In nutrient-poor sandy savannah soils in South Africa, Rughöft et al. [40] found a bacterial community dominated by Actinobacteria, Chloroflexi and Firmicutes members, except for Proteobacteria which were absent whereas they were well represented in our soils. In the sand dunes and inter-dune zones of the hyper-arid central Namib Desert, Ronca et al. [65] reported a similar bacterial community composition with seven phyla whose relative abundances were greater than 1% including Proteobacteria, Actinobacteria, Firmicutes and Chloroflexi members. The composition of the bacterial communities found in our study as regards the predominant phyla was clearly different to the composition of the bacterial community usually observed in temperate ecosystems and relatively similar to the composition of soil bacterial communities observed in soils from arid and semi-arid environments in several contexts like desert [35] [36], a land subsidence zone [66], an agroecosystem [67] [68], and an experimental precipitation gradient [69].
Senegal straddles the Sahel and experienced long drought events in the 1970s and 1980s [70] [71] with a dramatic decline in rainfall [72] and persistent dry conditions since then [19]. The climate of the region is also characterized by marked seasonality with a short rainy season in boreal summer, less rain in the winter season and high inter-annual variability [73]. Frequent extreme drying–rewetting events may select for microbial taxa that are more tolerant to desiccation stress, and these changes may result in a community that responds differently to moisture stress [74]. In a desiccation and rewetting experiment on Mediterranean grassland soils, Barnard et al. [75] pointed out that Proteobacteria, Chloroflexi, Firmicutes, Gemmatimonadetes and Planctomycetes members displayed stable and resistant life-strategies to response to environmental conditions.
Evans et al. [74] showed in soils exposed to frequent drying rewetting stress a bacterial community dominated by a greater proportion of stress tolerant taxa while in soils exposed to Ambient precipitation regime pulse- or drought-sensitive organisms predominate. Arid climates drive the assemblage of a community less sensitive to limited precipitation and adapted to desiccation stress by promoting the occurrence of more oligotrophic bacteria [38] [75]. According to Makhalanyane et al. [76] some Firmicutes spp. can form endospores that facilitate survival underdesiccating conditions. Members of Chloroflexi, Gemmatimonadetes and Verrucomicrobia that are considered to be less abundant in soils [64] have frequently been found in many dryland ecosystems while Gemmatimonadetes are known to be particularly adapted to low-moisture environments [77]. In our study, Acidobacteria and Bacteroidetes were present at relatively low abundance. Poor nutrient contents of our soils may explain the low relative abundance of Bacteroidetes members known to be copiotrophs [78] while the soil pH of our soils close to neutral might have impacted Acidobacteria members well adapted to low carbon availability [78] and known to increase in relative abundance when soil pH declines [45] [79].
4.2. Bacterial α and β-Diversity
Composition of the soil bacterial communities across the four pedoclimatic zones along the precipitation gradient revealed significant dissimilarities as shown by the Non-metric multi-dimensional scaling clustering of the soil samples according to their pedoclimatic origin (Figure 6(a)). Previous studies have shown that a change in environmental conditions, including rainfall gradient, can affect the composition of the community of soil bacteria [38] [48] [80] [81]. Structuring patterns of the bacterial β-diversity along the pedoclimatic gradient were predicted by shifts in the distribution of some OTUs mainly belonging to Bacillus, Rubrobacter, Pelobacter, Microvirga, Ktedonobacter, Conexibacter, Kaistobacter, Solirubrobacter, and Sphingomonas genera. Among the most abundant, these bacterial taxa showed spatial variations of their abundance along the gradient of environmental conditions (Supplementary Figures S1-S5) with divergent distribution patterns across the pedoclimatic zones even for taxa belonging to the same class or phylum. As Evans et al. [82] explain, climate-driven shifts in community composition could be due to the ecological traits of certain taxa that increase their ability to thrive under a particular climate regime. Bacterial assemblages exhibited vast physiological diversity at high taxonomical levels (class or phylum) and therefore spatial patterns of bacteria are more noticeable at low taxonomical levels (strain or species) because in this case ecological traits are common to the most members of the selected operational unit with consequently similar responses to environmental gradients [83]. Also, as said by Makhalanyane et al. [76], not all members of a given phylum are necessarily in the same ecological category and given the diversity present in some bacterial phyla, it is unlikely that an entire phylum shares the same ecological characteristics [78] [84]. These divergent responses probably contributed to make less perceptible changes in the community composition at higher taxonomic ranks like phylum or class. The exploration of changes in the community composition was more relevant at low taxonomic level. However, it is difficult to draw robust conclusions about the ecological properties of soil bacteria at that low level because well-characterized groups of bacteria generally concerned high taxonomic ranks such as phylum and class.
While bacterial community composition varied among the pedoclimatic zones, α-diversity did not vary across the pedoclimatic gradient (Table 3). It remained stable across the different pedoclimatic zones with a common pattern of diversity characterized by predominant taxa in the same proportions versus a lot of minority taxa. These findings underline the presence of dominant species along the gradient and confirm the observed opposing shifts in the relative abundance of some bacterial taxa that certainly contributed to the stability of the bacterial α-diversity across the pedoclimatic zones. Changes in microbial community structure do not necessarily lead to altered diversities, because changes in some taxonomic groups may be offset by changes in others [85]. Similar stability of the bacterial α-diversity in different soil water conditions has also been found in many other studies conducted in arid and semi-arid areas [48] [86]. Along a precipitation and temperature gradient in Eastern Inner Mongolia, Yaoet al. [38] found no significant differences in the observed species and bacterial α-diversity indices along a gradient of increased precipitation.
4.3. Relations between Bacterial Diversity and Environmental Parameters along the Rainfall Gradient
It is well known that environmental factors, including climate, soil type, soil properties and geographic location can affect soil microbial communities [66]. Soil pH, soil moisture and soil clay content were significantly correlated with changes in the bacterial community composition in our study (Figure 8). Influence of parameters for which we have observed a significant spatial variation along the pedoclimatic gradient suggest to a certain extent involvement of deterministic processes in the composition of soil bacterial communities.
Our study, like other studies in various ecosystems like cold desert [33] [87], lake sediments [88], a continental environmental gradient [45] [89], an elevation gradient [90], forests [91], deserts [92], wetlands [93], land use [67] [68] [94], and arable soil [95], reveals that pH is one of the most important driver of bacterial community diversity and composition. The db-RDA plot (Figure 8) clearly displayed this influence of soil pH, by allowing the separation of soils with a pH close to the neutrality from soils with a slightly more acidic pH. Like soil pH, spatial variations of soil moisture, result mainly from natural gradients or global climate change [48] and highly dependent on precipitation patterns [96], have significantly impacted bacterial β-diversity. Changes in microbial community composition and shifts in soil moisture due to variation in precipitation have been already report by some studies [39] [48]. Variation in soil moisture has a direct effect on bacterial communities due to the physiological stress experienced by these microorganisms, but also an indirect effect by regulating the availability of nutrients, which selects the best adapted bacteria to the soil conditions [47]. In this semi-arid region, soil moisture appeared also as for aboveground ecosystems, a strong predictor of belowground communities like soil microorganisms.
Soil clay content was the last significant factor correlated with changes in the composition of the bacterial community in our study. Soils of the region are characterized by a large mineral fraction, predominantly sandy with low levels of organic matter [97] [98], so soil clay content represents undoubtedly an important factor, determining soil texture and consequently soil moisture and nutrient contents. Soil clay content was positively correlated with soil moisture and soil nutrient content (Figure 2). In a study in the “Ferlo” region in Senegal, Fayeet al. [99] found a strong correlation between the soil moisture index and the nature of the geological formations, and stated that the variability of soil moisture was related to soil characteristics. Soil texture is important in determining soil moisture and nutrient status, which may have a significant impact on bacterial communities [100]. Xue et al. [49] determined that the effects of clay content and soil moisture content were considerable in controlling soil microbe variation along a climatic gradient. Hemkemeyer et al. [101] reported that bacteria can exhibit distinct preferences for a particular soil particle size fraction probably contributing to the spatial heterogeneity and bacterial diversity found in soils.
Soil pH, moisture content, and plant-derived organic carbon inputs are major factors known to shape the composition of soil microbial communities [3]. In our study, only soil pH and moisture content shaped the structure of the soil bacterial communities. This lack of organic matter impact may be due to the low level of organic matter that characterizes all the soil samples in a region where soils are known to be nutrient-poor. Soils are physically, chemically, and biologically heterogeneous, providing a wide range of niches to maintain microbial diversity [102]. Further studies on a variety of soils and environments are needed to obtain a more comprehensive picture of microbial community diversity and structure [103] in the semi-arid Sahelo-Sudanian region of Senegal.
5. Conclusion
The objective of this study was to explore in the Sudano-sahelian region of Senegal, the diversity and composition of soil bacterial communities along a pedoclimatic gradient and to improve our knowledge of the environmental factors that drive the spatial distribution of soil the bacterial community. Our study showed that the composition of the soil bacterial communities closely resembles that of arid to semi-arid environments with the dominant bacterial phyla in the soils being Firmicutes, Actinobacteria, Proteobacteria, Chloroflexi, Gemmatimonadetes, Acidobacteria, and Verrucomicrobia. We showed that, in the range of soil conditions created by climatic variations and soil heterogeneity, bacterial α-diversity was stable with a similar diversity pattern, while β-diversity highlighted different bacterial community structures as a function of the pedoclimatic origin of the soil samples. Our analyses revealed that the β-diversity variations were mainly driven by a few bacterial genera. In this semi-arid environment where annual mean rainfall is a determining factor in the type of land cover, soil moisture emerged as a strong predictor, and soil pH as the main environmental driver affecting the composition and the spatial distribution of bacterial communities in the Sahelo-Sudanian region.
Acknowledgements
We would like to thank Dr Laetitia Bernard (Eco & Sols), coordinator of the CAMMiSolE project. We also thank Dr Ranjard and his team at INRA UMR1347 Agroécologie, Dijon for the assistance and advices with R packages analysis. We also thank the LEMSAT Laboratory team L. Dieng, M. Gueye, M. Sané, L. Sagna, O. Faye (IRD, LMI IESOL, Senegal) for assisting with soil analyses and other laboratory work.
Funding
The present study was funded by the French Foundation for Research on Biodiversity (FRB-AAP-SCEN-2013 II—CAMMiSolE project). S.B. Diatta was funded by a grant from the CAMMiSolE project.
Supplementary Data
![]()
Table S1. Ecological characteristics and sampling details of the sites.
![]()
Table S2. 16S rRNA sequencing results. Number of OTUs and bacterial sequences by community fraction.
![]()
Table S3. Details of the Redundancy analysis (RDA) on the structure of the bacterial communities (Bray-Curtis dissimilarities) vs the environmental variables.
Permutation test for the evaluation of the overall significance of the fitted model.
Permutation test for the evaluation of the individual terms of the model retained by forward selection.
Biplot scores of explanatory variables on RDA axis.
![]()
Supplementary Figure 1. Spatial distribution of relative abundance of top 10 bacterial taxa within Actinobacteria class at the genus level along the pedo-climatic gradient. 1 to 4: Sahelian-sandy zone; 5 to 13: Sahelo-sudanian sandy zone; 14 to 18: Sahelo-sudanian loamy-sand zone; 19 to 25: Sudanian sandy-loam zone. Bacterial abundances are colored according to bacterial genera.
![]()
Supplementary Figure 2. Spatial distribution of relative abundance of top 10 bacterial taxa within a-proteobacteria class at the genus level along the pedo-climatic gradient. 1 to 4: Sahelian-sandy zone; 5 to 13: Sahelo-sudanian sandy zone; 14 to 18: Sahelo-sudanian loamy-sand zone; 19 to 25: Sudanian sandy-loam zone. Bacterial abundances are colored according to bacterial genera.
![]()
Supplementary Figure 3. Spatial distribution of relative abundance of top 10 bacterial taxa within Deltaproteobacteria class at the genus level along the pedo-climatic gradient. 1 to 4: Sahelian-sandy zone; 5 to 13: Sahelo-sudanian sandy zone; 14 to 18: Sahelo-sudanian loamy-sand zone; 19 to 25: Sudanian sandy-loam zone. Bacterial abundances are colored according to bacterial genera.
![]()
Supplementary Figure 4. Spatial distribution of relative abundance of top 10 bacterial taxa within Ktedonobacteria class at the genus level along the pedo-climatic gradient. 1 to 4: Sahelian-sandy zone; 5 to 13: Sahelo-sudanian sandy zone; 14 to 18: Sahelo-sudanian loamy-sand zone; 19 to 25: Sudanian sandy-loam zone. Bacterial abundances are colored according to bacterial genera.
![]()
Supplementary Figure 5. Spatial distribution of relative abundance of top 10 bacterial taxa within Bacilli class at the genus level along the pedo-climatic gradient. 1 to 4: Sahelian-sandy zone; 5 to 13: Sahelo-sudanian sandy zone; 14 to 18: Sahelo-sudanian loamy-sand zone; 19 to 25: Sudanian sandy-loam zone. Bacterial abundances are colored according to bacterial genera.