Electrochemical Biosensor for Multiple Methylation-Locus Analysis Based on DNA-AuNPs and Bienzymatic Dual Signal Amplifications ()
1. Introduction
DNA methylation has always been the cutting-edge of epigenetic studies, which plays a significant role in cellular development, genomic stability, gene expression and regulation. In eukaryotes, DNA methylation occurs mainly at the carbon-5 position of cytosine residues at CpG dinucleotides, resulting in the formation of the 5-methylcytosine (5-mC). Normally, a majority of CpG regions in the promoter regions of tumor suppressor genes are unmethylated, once the aberrant methylation occurs, these genes are transcriptionally repressed and inactivated, which is considered as an early warning of tumorigenesis [1] - [8]. Therefore, the determination of DNA methylation can benefit the assessment and classification of tumors. Traditional DNA methylation assays, such as bisulfite sequencing (BS-seq), restriction endonucleases (RE) analysis, high-performance liquid chromatography (HPLC), mass spectrometry (MS), DNA methylation microarrays [9] - [13], generally demand time-consuming, intricate processes or expensive large-scale instruments, which extremely limit their utilization in the analytical and clinical applications. However, electrochemical biosensors offer superior features owing to its simplicity, portability, sensitivity and fast response [14] [15] [16]. Up to now, a number of electrochemical strategies have been established for the detection of DNA methylation. For instance, Geng and Bao utilized the DNA methyltransferases (DNA MTase) to catalyze the formation of 5-mC coupled with the specific cleavage of methylation-sensitive endonucleases HpaII, which could cleave the unmethylated DNA and lead to the decrease of electrochemical signals of methylene blue (MB) to obtain the methyltransferase detection limit of 0.04 U/mL with the DNA concentration of 1 μM [17]. Using 5-mC antibody, Wang et al. built an electrochemical biosensor which can easily detect the MTase activity and DNA methylation with a detection limit of 0.5 μM [18]. Xu et al. also presented a MTase activity detection approach based on the signal amplification of enzyme-modified on the gold nanoparticles to obtain a lower detection limit of 0.17 pM [19]. However, most of reports achieved DNA methylation levels through the enzymatic reactions conducted by DNA MTase without the accurate detection of methylation-locus directly.
As an ideal methylation-specific recognition molecule, the methyl-CpG binding domain (MBD) proteins could be utilized for the quantitative analysis of methylation patterns due to its superior methylation-loci specificity. It is reported that the MBDs include at least the MeCP2, MBD1, MBD2 and MBD4. They are capable of binding methylation-locus in double-stranded through their functional MBD domain, indicating that MBDs could be used to serve as the specific recognition unit for the detection of DNA methylation-locus [20] [21].
Immunoassay exhibits high specificity and sensitivity in which the employed immunomolecules were labeled with certain enzyme as molecular labels. Horseradish peroxidase (HRP) is known for its robust capability to catalyze various chemical oxidations involving hydrogen peroxide (H2O2), and it has been extensively employed in electrochemical immunoassays by transforming the redox reaction process into a detectable electrochemical signal [22] [23] [24]. Glucose oxidase (GOD) is also widely used as the signal tracer which specifically catalyzes the oxidation of glucose to gluconic acid, while O2 was reduced to H2O2 [25] [26]. Theoretically, these two enzymes could be co-labeled on the antibody molecule as amplifying labels. The GOD is catalytically linked to HRP to form bienzymatic cascade scheme, which could produce a further enhanced response signal rather than the one that labeled with mono-enzyme.
The purpose of this study, which involves the methylation-loci recognition unit of MeCP2 and dual signal amplification from powerful content of DNA anchoring gold nanoparticles (DNA-AuNPs) coupled with bienzyme labeled antibody, is to present an ultrasensitive electrochemical biosensing method for the quantitative analysis of multiple methylation-locus in particular sequence without the need for bisulfite treatment or gene amplification. The electrodeposited AuNPs could enlarge the specific surface area of the electrode, which effectively promote the electron transfer on the electrode and increase the anchoring sites for the self-assembling of the thiolated DNA probes via strong Au-S bond [27] [28] [29]. Thus, the DNA-AuNPs has the potential ability for the enhancement of sensitivity. Also, the bienzyme labeling system could conduct the robust cascade catalysis to produce a further amplified signal and therefore enhanced the sensitivity of the biosensor. As illustrated in Scheme 1, the thiolated single-stranded DNA probe was first self-assembled on the surface of the AuNPs-modified electrode. After the hybridization with the complementary oligonucleotides, the conjugation of the His-tagged MeCP2 was performed. Thereafter, the bienzyme labeled antibody binds to the His-tag of MeCP2 via immunoreactions. In the buffer solution containing glucose and hydroquinone, the GOD could catalyze the glucose substrate to generate H2O2 by the reduction of ambient oxygen. Then the in situ generated H2O2 could be rapidly consumed by the nearby HRP. The combination of GOD and HRP formed bienzymatic cascade catalysis to initiate the oxidation of hydroquinone, generating a detectable amplified oxidation current. The current signal was directly related to the amount of methylation-loci, therefore allowing the precise determination of multiple methylation-locus. Impressively, owing to the superior methylation-loci specificity of MeCP2 and dual signal amplification procedures, this designed method could be a reliable tool for the accurate detection of DNA methylation.
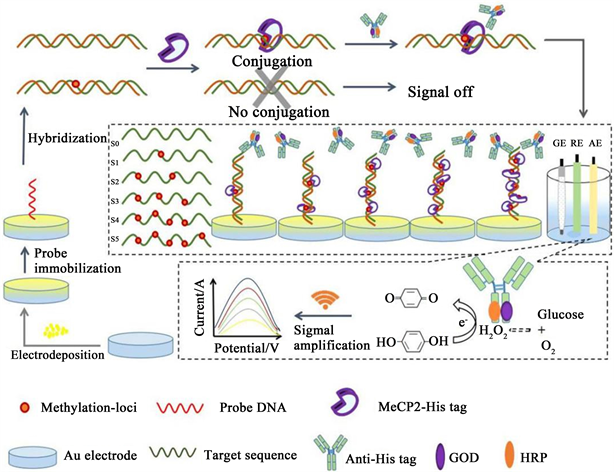
Scheme 1. Schematic illustration of the electrochemical biosensing method for the detection of DNA methylation.
2. Materials and Methods
2.1. Materials
Hydrogen tetrachloroaurate trihydrate (HAuCl4∙3H2O), Tris-EDTA (TE, pH 7.4) buffer solution and tris (2-carboxyethyl) phosphine (TCEP) were obtained from BBI Life Science Corporation. Bovine serum album (BSA) was purchased from Shanghai Solarbio Bioscience & Technology Co. Ltd., Recombinant human methyl-CpG-binding protein 2 (MeCP2) was from novoprotein. MeCP2 was produced by mammalian expression system and the target gene encoding Met1-Ser486 was expressed with 6 × His tag at the C-terminus. Anti-6 × His tag mouse monoclonal antibody, GOD and HRP were obtained from Shanghai Sangon Biotechnology Co. Ltd. The oligonucleotide sequences (Table 1) were synthesized and purified by Shanghai Sangon Biolotechnology Co. Ltd. (China). The oligonucletides were dissolved in TE buffer and stored at −20˚C.
The buffer solutions used in this work were prepared as follows: Probe immobilization buffer: 1 × TE buffer (pH 7.4) containing 50 mM NaCl and 1.0 mM TCEP. DNA hybridization buffer: 1 × TE buffer (pH 7.4) containing 50 mM NaCl and 10 mM MgCl2. Phosphate buffered saline (PBS, pH 7.0): 0.01 M NaH2PO4, 0.01 M Na2HPO4 and 0.01 M NaCl. MeCP2 protein immobilization buffer: 0.1 M phosphate buffered saline and 5% glycerol. Other reagents were of analytical grade and all the solutions were prepared using ultra-pure water (18.2 MΩ∙cm resistivity).
![]()
Table 1. Oligonucleotide sequences used in this work.
*5’ end of probe DNA S was modified with -HS, and the i5med represents the methylated cytosine base. The methylation site of the target DNA was located two bases away from the 5’ end, and the multiple methylation sites were separated randomly with several other bases.
All electrochemical measurements were performed on a CHI660D electrochemical workstation with a conventional three-electrode system: platinum wire as auxiliary electrode, Ag/AgCl electrode as reference electrode, and a gold electrode as working electrode. The morphology of nanoparticles and probe immobilization were estimated using scanning electron microscope (SEM, crossbeam 340 zeiss, Germany) and atomic force microscopy (AFM, IPC-208B, Chongqing University), respectively.
2.2. Methods
Preparation of bienzyme co-labeled antibody
The co-labeling of HRP and GOD on the anti-His tag antibody was dealt with a two-stage process. To begin with, the labeling of GOD was performed using the glutaraldehyde cross-link method [30] [31]. The reaction mixture consisted of 1 mg/mL antibodies and 2 mg GOD in 1.25% glutaraldehyde solution. Subsequently, the GOD labeled antibodies were coupled to HRP using the sodium periodate procedure through their carbohydrate moieties [32]. The activation of HRP was done in a solution containing 60 mM sodium periodate and 160 mM ethylene glycol. Exactly 1:1 molar rations of antibody to enzyme were taken for the labeling process. Followed by centrifugation and resuspending, the mixture was dialysed in 0.15 M PBS (pH 7.4) and stored at −20˚C. The obtained products were noted as GOD-HRP/anti-His tag.
Preparation of AuNPs modified electrode
The 2 mm diameter gold electrode was polished to a mirror-like surface with 0.05 μm alumina slurries, then the polished electrode was ultrasonicated in ethanol and ultrapure water for 5 min respectively. Next, the electrode was soaked in piranha solution (98% H2SO4:30% H2O2, 3:1, V/V) for 15 min. After activation, the bare gold electrode was subjected to electrochemical sweeping in 0.5 M H2SO4 until the cyclic voltammetry (CV) curve was stable and reduplicative. Followed by thoroughly rinsed with ultrapure water, 3 mM HAuCl4∙3H2O solution containing 0.1 M KNO3 was used for the potential deposition at −0.2 V for 120 s. Finally, the obtained AuNPs/Au was cleansed again and dried with nitrogen.
Probe immobilization and hybridization
The probe immobilization was conducted by dropping 10 μL of probe immobilization buffer solution (containing 0.1 μM thiol-capped probe DNA S) on the surface of AuNPs/Au and incubated for 12 h at 4˚C condition to form the Au-S bond. The obtained ssDNA/AuNPs/Au electrode was finally prepared for hybridization. Prior to hybridization, electrode was rinsed thoroughly with PBS and ultrapure water to remove the extra absorbed DNA probe. Then 10 μL hybridization buffer (containing 0.1 μM target DNA) was dropped on to the probe modified electrode for 60 min at 37˚C in a humid atmosphere.
MeCP2 protein immobilization and immunoreactions
After hybridization, the electrode was incubated with 0.5% BSA solution (prepared in 0.1 M PBS) for 30 min. Following that, 10 μL of MeCP2 protein (200 μg/mL) was further dropped and incubated for 60 min at 37˚C. Washed with PBS repeatedly, the electrode was incubated with 10 μL GOD-HRP/anti-His tag (10 μg/mL) at 37˚C. Afterward, the electrode was rinsed again with PBS.
Electrochemical characterization and determination
Cyclic voltammetry (CV) and electrochemical impedance spectroscopy (EIS) were used to characterize the assembled process of electrode in 3 mM [Fe(CN)6]3−/4− (1:1) solution containing 0.1 M KCl at a scan rate of 50 mV/s. To detect the electrochemical signals, differential pulse voltammetry (DPV) was carried out in 10 mL of 0.1 M PBS containing 0.5 mM glucose and 0.25 mM hydroquinone with the scan potential range from −0.1 V to 0.6 V. Other parameters were as follows: pulse width, 0.05 s; pulse amplitude, 50 mV; sampling width, 0.0167 s; pulse period, 0.5 s; quite time, 2 s.
3. Results and Discussion
UV-vis spectrophotometer analysis
The coupling products were identified by ultraviolet and visible (UV-vis) scanning at a range of 200 - 800 nm. As can be seen in Figure 1, two absorption peaks are observed at the wavelength of 403 nm and 275 nm, representing the UV absorption spectra of HRP/antibody and GOD/antibody, respectively [32] [33] [34]. Due to the combined modification of antibody with these two enzymes, there was shift of absorption peak at 403 nm and 275 nm, which confirmed an efficient bienzyme modification to the antibody.
Morphological characterization of AuNPs/Au and ssDNA/AuNPs/Au
According to the SEM image (Figure 2(a)), it can be observed that the AuNPs are spherical and in good distribution with an average particle size of 32 nm, so the superficial area of the electrode was enlarged. The AuNPs/Au and ssDNA/AuNPs/Au were compared by AFM. Figure 2(b) and Figure 2(c) show the AFM images of AuNPs monolayer and the immobilization of the captured
![]()
Figure 1. Image of UV absorption spectra of HRP/anti-His tag (a), GOD/anti-His tag (b) and GOD-HRP/anti-His tag (c).
![]()
Figure 2. (a) SEM image of AuNPs/Au. (b) AFM image of AuNPs/Au. (c) AFM image of ssDNA/AuNPs/Au.
probe. The AFM parameters have been evaluated for 1000 × 1000 nm and 2000 × 2000 nm surface area. It is significant that there are morphological differences between both the films. The view of AuNPs/Au (Figure 2(b)) shows uniformly deposited homogeneously dispersed AuNPs on electrode, which showed consistent particle sizes with those of SEM. After the modification of ssDNA probe, it can be observed the high brightness white dots which were marked by a white circle and the surface morphology became rougher (Figure 2(c)), giving the increased height of ssDNA anchoring on the surface of the AuNPs/Au films.
Electrochemical characterization of the biosensor
EIS was employed to characterize the electrode fabrication process. As depicted in Figure 3(a), curve a shows a small semicircle, which indicates a very low impedance of the bare gold electrode. When the bare gold electrode was modified with AuNPs, a straight line was observed (curve b), suggesting that the AuNPs could effectively promote the electron transfer of [Fe(CN)6]3−/4−. However, since the negatively charged phosphate backbone of DNA could repulse the diffusion of [Fe(CN)6]3−/4− on the electrode surface, the impedance value increased after the probe DNA was immobilized on the AuNPs/Au surface (curve c), and a further increase was observed after the hybridization process (curve d). After the MeCP2 protein conjugated to the DNA methylation-loci, the assembled protein could also block the transfer on the electrode, so the impedance value continued to elevate (curve e).
We also employed CV to monitor each step of the fabricating process (Figure 3(b)). The CV curve showed a pair of distinct redox peaks, indicating that the electron transfer of [Fe(CN)6]3−/4− on the electrode surface. The bare gold showed a well-defined redox peak with the current of 0.625 μA (curve a). The separation of peak-to-peak (ΔEp) was 0.077 V. After the AuNPs were electrodeposited on the bare gold surface, the redox peak current increased with the decreased ΔEp (curve b). This phenomenon was caused by the excellent conductivity of AuNPs, which increased the electrode effective surface and improved the electron transfer
![]()
Figure 3. Characterization of EIS (a) and CV (b) obtained for the fabricated process. a. Bare gold electrode; b. AuNPs modified gold electrode; c. Probes self-assembly; d. Hybridization with target DNA; e. MeCP2 conjugation.
rate. However, the redox peak current of [Fe(CN)6]3−/4− decreased when the DNA probe was immobilized through the Au-S bond due to the electrostatic repulsion force between the negatively charged [Fe(CN)6]3−/4− and the negatively charged DNA (curve c), thereby decreasing the redox peak current and enlarging the distance between two peaks. In short, the electrode surface had been successfully modified with this probe. Then, the redox peak current further decreased after the probe DNA hybridized with target sequences (curve d) due to the number of negative charges on the gold electrode surface continued to rise, so the electron transfer ability of [Fe(CN)6]3−/4− was further attenuated, which then reduced the redox peak current. Subsequently, when MeCP2 protein conjugated with the methylated DNA, the redox peak current decreased obviously (curve e). The current decrease can be attributed to the steric hindrance caused by the large volume of MeCP2 protein, which blocked the diffusion of [Fe(CN)6]3−/4− towards electrode surface and decreased the electron transfer rate. Collectively, each modification step of the electrochemical biosensor was successful. This was in accordance with the results of other studies [18] [19]. The CV results correspond to the EIS for each step of the electrode modification process, which confirmed the successful modification steps on the working electrode.
Optimization of experimental conditions
Optimized experimental parameters including the hybridization time, MeCP2 and substrates concentration are vital to the performance of the proposed method. All the parameters mentioned above were validated and we selected the optimal experimental conditions according to the results. Parameters of 60 min (hybridization time), 200 μg/mL (MeCP2 concentration), 0.5 mM (glucose substrate concentration) and 0.25 mM (hydroquinone substrate concentration) were used in the study.
Analysis of DNA methylation patterns in particular sequence
DNA S0-S5 containing known amount of methylation-locus were hybridized with probe DNA S, and then the MeCP2 and antibody conjugations were validated. Figure 4(a) shows the DPV results derived from the target sequence with various methylation-locus and the peak current increased with the growing numbers of methylation-locus. Due to the quantitative conjugation of the MeCP2 to the methylation-loci in the target sequence, the subsequent specific recognizing by the bienzyme modified antibodies, and the current signal generations under the catalysis of the enzymes of GOD and HRP, the current signal were highly related to the levels of the DNA methylation-locus and the linear relationship was exhibited in Figure 4(b). On the contrary, a weak background current for the unmethylated sequence is also observed (Figure 4(a), curve a) for there was only a small amount of nonspecifically absorbed proteins or antibodies on the electrode. The results demonstrated that the proposed strategy can be applied to the selective identification of DNA methylation patterns.
Characterization of the bienzyme signal amplification
In order to prove the electrocatalytic effect of the bienzyme system, we conducted a comparison test by using three kinds of labeled antibodies with GOD/anti-His tag, HRP/anti-His tag and GOD-HRP/anti-His tag, respectively. The tests were investigated through the DPV signal to 0.1 μM target sequence with one methyaltion-loci. As shown in Figure 5, the bienzyme system shows a strong oxidation current of about 6.37 μA (curve c), increased to 3.2 fold than the monoenzyme HRP (curve b). In contrast, an extremely low current signal was obtained when the anti-His tag antibody was merely modified with GOD (curve a), indicating that the oxidation of hydroquinone by H2O2 was inefficient without HRP. These results indicate that the bienzyme co-labeling procedure was successful and could considerably enhance the sensitivity of the biosensor.
Sensitivity, repeatability and stability of the biosensor
We evaluated the DPV responses of different DNA concentrations with only one methylation-loci. Figure 6(a) indicates that the peak currents are increasing along with increased concentration of target DNA across the range of 10−15 M to 10−7 M, and the linear relationship between DPV peak current and the logarithm
![]()
Figure 4. (a) DPV analysis of methylation patterns S0-S5 (a-f), DNA S0 was unmethylated. DNA S1-S5 have 1, 2, 3, 4, 5 methylation locus in each sequence. (b) The linear relationship between peak current and the amount of methylation-locus from 1 to 5.
![]()
Figure 5. DPV responses obtained for the different enzyme-labeled antibodies. (a) GOD labeled anti-His tag. (b) HRP labeled anti-His tag. (c) GOD and HRP co-labeled anti-His tag.
of the DNA concentration can be expressed as I (μA) = 0.548 log C (M) + 10.306 with a correlation coefficient (R) of 0.989 (Figure 6(a)). Comparing to the previous assays (Table 2), our method is proven to be a sensitive tool for the detection of DNA methyaltion.
The repeatability of the proposed method was conducted by detecting the target DNA with one methylation-loci for five repeat measurements. The relative standard deviation (RSD) value was 4.6%, which is lower than previously reported studies [35] [36] [37]. The results illustrate an ideal repeatability of the proposed method.
The long-term stability of the biosensor was also investigated. We stored the prepared biosensors in pH 7.4 PBS at 4˚C and detected every seven days. The current responses were stable, and finally retained 93.7% of its initial redox current after storage of 28 days.
4. Conclusion
In this work, we successfully fabricated an enhanced electrochemical biosensor for the quantitative analysis of multiple methylation-locus in particular sequence with MeCP2 as the specific recognition unit. A lower detection limit of 0.1 fM was achieved owing to the dual signal amplification of the AuNPs and bienzymatic
![]()
Figure 6. (a) DPV responses of different DNA concentration (a - k: 10 - 17 M, 10 - 16 M, 10 - 15 M, 10 - 14 M, 10 - 13 M, 10 - 12 M, 10 - 11 M, 10 - 10 M, 10 - 9 M, 10 - 8 M, 10 - 7 M, respectively). (b) The linear relationship of peak current versus the logarithm of DNA concentration.
![]()
Table 2. Comparison with previous reports.
cascade catalysis. The repeatability and stability were also validated. Comparing with other biosensing approaches based on conventional bisulfate conversion or methylation restriction enzyme cleavage, our presented method is much easier, cheaper and more time-saving, indicating that our method possesses great prospect for the clinic application in the early diagnosis of cancer and other methylation-related diseases.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (No. 81572078, No. 81401722 and No. 81873982), the Projects of Basic Research and Frontier Exploration in Chongqing (No. cstc2019jcyj-zdxm0037), and the army Innovation Research Project (No. LJ20182B060005).