Automatic Sleep Spindle Detection with EEG Based on Complex Demodulation Method and Decision Tree Model ()
1. Introduction
Sleep is an important physiological activity of human being. According to R & K criteria [1], sleep is described with awake, non-rapid eye movement (NREM) and rapid eye movement (REM). NREM is further divided into four stages: Stage 1 (S1), Stage 2 (S2), Stage 3 (S3) and Stage 4 (S4). Sleep spindle is the characteristic waveform of sleep stage 2 [2]. It is a transient waveform with waxing- waning amplitude. The frequency of sleep spindle is about 12 - 14 Hz, and its duration should be at least 0.5 seconds [1]. Sleep spindle is related to the investigation of sleep [3]. In addition, the characters of spindle such as power [4] and density [5] are related to cognition and memory of human.
In clinics, qualified clinicians make visual inspection on sleep spindles [6]. Usually, variable shapes of waveform may appear in EEG (electroencephalogram). The inspection requires qualified skill and clinical experience. The visual inspection is rather laborious, time-consuming and subjective. In order to improve the efficiency of sleep spindle detection, several automatic methods had been developed including signal processing by time-frequency methods and decision making by thresholds or machine learning techniques. Time-frequency methods include Teager Energy Operator (TEO) [7], Continues Wavelet Transform (CWT) [8], and Matching Pursuit (MP) [9]. It is easy to obtain the energy of signal within a certain frequency band, but the result of TEO was sensitive to noise [10]. CWT can meet the time frequency resolution only if the “mother wavelet” is appropriate. MP is good at signal decomposition, but this advantage is based on the assumption that the “atom of dictionary” can well represent the signal components, which is difficult to promise in real data. In another hand, it is convenient to apply thresholds to detect sleep spindle. However, it largely depends on the experience of experts. Furthermore, it is difficult to account for all subjects [11]. Compared to the defined thresholds, machine learning techniques such as Support Vector Machine (SVM) or Artificial Neural Network (ANN) [12] are more objective. Though both SVM and ANN can well detect sleep spindle, they cannot describe the various shape of sleep spindle. Therefore, it is still necessary to develop the automatic method to detect sleep spindle, which can capture the transient signal, describe the characteristics and account for the variability.
In this paper, the complex demodulation method (CDM) and decision tree model are adopted to detect sleep spindle automatically. The sleep data of EEG were recorded based on PSG (polysomnogram) measurement. Firstly, the data segments are preliminarily analyzed to extract necessary segments from the continuous sleep EEG recording. Secondly, according to the time-frequency character and instantaneity of sleep spindle, CDM is adopted to extract the candidate waveforms and calculate several features. A decision tree model is constructed based on a set of training samples. Final judgments were obtained for the test samples. Furthermore, the detected results were used to amend the sleep stage recognition of S2 to evaluate the validity of sleep spindle detection.
2. Method
2.1. Data Acquisition
The overnight PSG recordings were obtained from 5 subjects. Detail explanation was done for all the subjects before recordings and informed consent was obtained. The PSG measurement includes 4 EEG channels (C3/A2, C4/A1, O1/A2, O2/A1), 2 EOG (electrooculography) channels (LOC/A1, ROC/A1) and 1 chin EMG (electromyography) channel (chin-EMG). In this study, EEGs were analyzed for sleep spindle detection recorded under a sampling rate of 100 Hz, with a high frequency cut off of 35 Hz and a time constant of 0.3s. The recorded data were segmented into 30-second epochs. The consecutive epochs were inspected according to R & K criteria by a qualified clinician.
2.2. Pre-Processing
Sleep spindle is mainly related to light sleep. It is not necessary to analyze all the recorded data. There are two parameters calculated in Table 1. The ration of α component is calculated with EEG in O1/A2 and O2/A1 channels, high frequency component is in
C3/A2 and C4/A1 channels. If Rα is larger than 0.25 or Rh is larger than 0.025, the epoch is excluded for sleep spindle detection.
The remaining data is normalized by Z-score so that the difference of amplitude of EEG among subjects can be minimized. X0(n∆t) represents EEG signal and X(n∆t) represents the result of normalization,
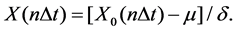 (1)
(1)
In Equation (1), n = 1, 2,∙∙∙, N, μ and δ represents the average and the standard deviation of X0(n∆t).
2.3. Complex Demodulation
CDM is a time-frequency domain method of transient signal processing. The change of signal amplitude over time can be obtained as long as the frequency of the signal is given. A time series X(n∆t) can be represented as,
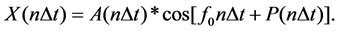 (2)
(2)
In Equation (2), A(n∆t ) is a changing amplitude and P(n∆t) is a changing phase. A(n∆t) can be extracted as follows if the frequency f0 is known. Shift all the frequency in X(n∆t) by - f0,
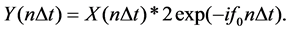 (3)
(3)
Y(n∆t) in Equation (3) can also be represented as,
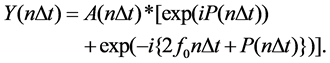 (4)
(4)
Let Y’(n∆t) be the complex signal when Y(n∆t) is passed through a low-pass filter,
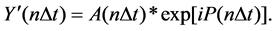 (5)
(5)
![]()
Table 1. Parameters for pre-processing.
Therefore, A(n∆t) can be obtained as,
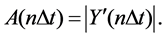 (6)
(6)
In this study, EEG signals in channel C3/A2 and C4/A1 were analyzed by CDM for every 30s epoch after pre-processing. X(n∆t) is the normalized EEG data. The instantaneous amplitude Aθ(n∆t) of θ wave (4 - 7 Hz), Aα(n∆t) of α wave (8 - 13 Hz), and Aspindle(n∆t) of spindle (12 - 14 Hz) are obtained when f0 in the above equations is set as 6 Hz, 10 Hz and 13 Hz correspondingly. Here, Aspindle(n∆t) is the main information for candidate sleep spindle detection and feature calculation. Aθ(n∆t) and Aα(n∆t) are used as additional information for feature calculation.
According to the various of Aspindle(n∆t) with time, if the instantaneous amplitude of one local maximum point with CDM is larger than 0.5 μv, a peak point tp is marked. The starting and ending points of t1 and t2 are the position of the nearest points from the left and right of tp whose instantaneous amplitude is smaller than half of Aspindle(tp). The corresponding EEG signal is selected as candidate waveform.
2.4. Decision Tree Model
For the candidate waveform after CDM processing, four parameters in Table 2 are calculated. The parameter of peak is defined as the maximum instantaneous amplitude of candidate wave around 13 Hz, while slope is the average changing rate of the wave. R1 is the average amplitude ratio of spindle and θ wave, while R2 the ratio of spindle and α wave.
A set of sleep spindles is adopted as the training data. The values of parameters are divided into 3 levels: “high”, “middle”, “low”. After training, a decision tree model is constructed to recognize the sleep spindle for the test data.
3. Results
3.1. Decision Tree Model
After pre-processing and CDM analysis, candidate sleep spindles were obtained. The 400 samples from 2 healthy subjects were adopted as the training data. The candidate waveforms detected form 3 patients were for test.
The obtained decision tree model was illustrated in Figure 1, square represents attribute node and circle represents leaf node while “T” means spindle and “F” means non-spindle. Label “0”, “1”, “2”, “0/1”, “1/2” correspond to
![]()
Table 2. Parameters for sleep spindle detection.
![]()
Figure 1. Decision tree model for sleep spindle recognition.
level “low”, “middle”, “high”, “low or middle” and “middle or high”. According to the obtained model, it can be summarized that parameter of peak was the most important feature for sleep spindle detection and followed by R1, R2 and slope successively.
3.2. Sleep Spindle Detection
The process of automatic sleep spindle detection was shown in Figure 2. The raw EEG signals of three samples were illustrated in Figure 2(a). The sleep spindle in (a − 1) is quite clear to inspect. However, the sleep spindle in (a − 2) is a sleep spindle mixed with θ component (4 - 7 Hz), while (a − 3) is a sleep spindle with smaller amplitude. Figure 2(b) were the results of CDM, in which X-axis is time and Y-axis is Aspindle(n∆t). The Aspindle(n∆t) curves contain a peak, which means the amplitude of EEG signals around 13Hz went through the process of waxing and waning. The peak points tp, the starting points t1 and ending points t2 were marked by circles in Figure 2(b). Comparing the raw data and CDM results, the obtained Aspindle(n∆t) curves indicated the trend of amplitude around frequency of sleep spindle clearly.
Parameters were calculated and evaluated according to Table 2. All the parameters of standard sleep spindle in (a − 1) were “high”. The amplitude trend of signal around 13 Hz was obvious and the shape of the signal was concentrating. For sleep spindle in (a − 2), R1 was “low”, while other parameters were “high”. Although the shape of wave was less concentrating, it was considered as sleep spindle. For sleep spindle in (a − 3), its peak was “low” and slope was “middle”, while both R1 and R2 were “high”. The changing of amplitude was slower. However, the shape of the wave was concentrating so that it was regarded as sleep spindle. In the sleep EEG data of patients, most of the shapes of sleep spindle may differ from the typical ones. The constructed decision tree model based on CDM analysis interpreted the sleep spindles in real cases.
3.3. Amendment to Sleep Stage 2
The overnight sleep data of 3 subjects were analyzed by visual inspection and automatic sleep stage determination [13]. Sleep stage 2 is usually inspected by
![]()
Figure 2. Sleep EEG data analysis by complex demodulation for sleep spindle detection.
![]()
Table 3. Sleep spindle detection for sleep stage 2 recognition.
characteristic waveforms. Due to the instantaneous character of the waveforms, it is rather difficult to extract valuable parameter which can be included into the feature vector for automatic sleep stage recognition algorithms. It would affect the recognition accuracy of S2 as well as the total accuracy. In this study, the effectiveness of sleep spindle detection by using the presented method was evaluated as amendment for S2.
In Table 3, “A” represented the automatic sleep stage recognition result while “B” showed the improvements by using automatic sleep spindle detection. For subject 1, there were 15 epochs can be amended for the false-negative epochs. The recognition accuracy improved from 73% to 76%. The same effectiveness can be observed from another two subjects from 63% to 87% and 81% to 90% respectively. In another hand, there were no epochs detected with sleep spindle for false-positive ones.
4. Discussion and Conclusions
Sleep spindle is a kind of characteristic waveform which has instantaneous character mainly appeared in sleep stage 2. According to the definition of sleep spindle, it has obvious frequency characteristic. However, the frequency characteristic is not enough to make the judgment. The shape of real sleep spindles in sleep EEG data may be various and mixed with the background EEG. Those increased the difficulty for automatic detection.
In this study, CDM is adopted for the detection of candidate waveforms which has similar frequency characteristics with sleep spindle. A decision tree model is constructed according to the features calculated based CDM. The obtained model showed similar manner to the visual inspection which can be adaptive to the various shape of sleep spindles in real clinical data. The detection results increased the recognition accuracy of sleep stage 2 among the subjects. The developed technique is useful for sleep spindle detection, which can also be combined with various sleep stage recognition method as an amendment tool.
Acknowledgements
This research was financially supported by the Medical Cooperation Project by Shanghai Municipal Science and Technology Commission 12DZ1940903, the Shanghai Natural Science Foundation under Grant 16ZR1407500.