1. Introduction
Biological control is an environmentally-friendly alternative to chemical pesticides and it is an attractive method protecting the plants from pathogens, because the wide usage of chemicals has a negative impact on the environment and human health. Many biocontrol agents were isolated by screening of the large number of soil or plant-associated microorganisms for antagonism against phytopathogens in vitro or in planta [1] [2] . A number of bacterial species with antagonistic activity were isolated from the rhizosphere of different plants. Among those, bacilli and pseudomonads are the most common isolates [1] [2] [3] [4] .
It is known that various species of the Bacillus genus are able to stimulate the plant growth [5] . Bacteria can promote plant growth through a number of mechanisms, such as improvement of plant nutrition; induction of systemic resistance; toxicity to pests and antagonism pathogens [5] [6] [7] . The antagonistic activity of Bacillus is associated with the synthesis of various antimicrobial peptides [8] [9] , secreted enzymes [10] , proteins [11] and volatile organic compounds (VOCs) [5] [10] . Many Bacillus isolates were shown to have antifungal activity against phytopathogenic fungi that make them good biocontrol candidates [12] [13] [14] . Cyclic lipopeptide fengycin plays an important role in this process [15] .
The strains of the species B. subtilis can vary considerably both phenotypically and genetically and that affects their antagonistic potential. Comparative analysis of proteomes of two B. subtilis strains with antagonistic potential highlighted the major differences in the composition of intracellular and extracellular proteins [11] [16] , some of which can be associated with antimicrobial properties. Because of their fast growth, ability to effectively grow in low cost media and to sporulate under undesirable conditions Bacillus isolates are the attractive candidates for application as the biocontrol agents. There is a growing demand for such agents since it is expected that global market for biopesticides will significantly expand in the next 3 - 5 years (www.bccresearch.com/market-research/chemicals/biopesticides-chm029e.html; [17] ).
The aim of this study was to directly compare two antagonistic strains of Вacillus spp. isolated from potato roots, in their ability to suppress phytopathogenic fungi through production of cyclic lipopeptides, hydrolytic enzymes and siderophores. In addition, we evaluated plant growth promoting potential of B. subtilis GM2 and GM5 on wheat seedlings. Finally, we showed that one of the two B. subtilis isolates, GM5, can protect wheat seedlings against Fusarium oxysporum and, therefore, represents a biocontrol candidate with the potential for its application in agriculture.
2. Material and Methods
2.1. Isolation and Identification of Antagonistic Strains
Several bacterial strains were isolated from the rhizosphere of potato roots. The non-rhizosphere soil was removed by gentle shaking. The rhizosphere-asso- ciated soil was collected from roots by dipping and gentle shaking in sterile water under aseptic conditions. The resulting soil suspension was used to inoculate LB agar plates and pure cultures were obtained by repetitive streaking to single colonies on the additional LB plates. Plates were incubated for 48 h at 28˚C. For long-term storage bacterial strains were kept at ?70˚C in LB-broth with 15% glycerol (v/v).
The colonies with different colony morphologies were selected for further studies. Bacterial isolates were screened for their activity against Fusarium sp. as described in section In vitro antagonistic activity. Two bacterial isolates with the highest inhibitory activity were selected for further characterization.
Pure bacterial cultures grown overnight at 30˚C were analyzed by MALDI BioТyper (Bruker Daltonik). This technique is based on the comparison of a number of bacterial proteins against preexisting database. Scores were calculated by Biotyper software as arbitrary units with values between 0 and 3 as a result of comparison of each sample mass spectrum to the reference mass spectra in the Bruker database. Species identification is possible when the of score values lie between 2.300 and 3.000 according to manufacturer’s recommendations. Genomic DNA was isolated by standard technique [18] . The 16S rRNA genes were amplified by PCR using primers: Fw 5’-gagtttgatcctggctcag, Rev 5’-acggttacc- ttgttacgactt. DNA gyrase subunit B genes were amplified with the following primers: UP1 5’-GAAGTCATCATGACCGTTCTGCAYGCNGGNGGNAARTTY- GA and UP2r 5’-AGCAGGGTACGGATGTGCGAGCCRTCNACRTCNGCRT- CNGTCAT. PCR analysis was performed according to the established protocol [19] . PCR products were separated on agarose gel [18] .
The obtained sequences were analyzed using the BLAST software (https://blast.ncbi.nlm.nih.gov/Blast.cgi).
2.2. Phytopathogenic Fungi and Culture Condition
In this study we used the following phytopathogenic fungi: Fusarium avenaceum, Fusarium oxysporum, Fusarium redolens, Fusarium solani, Fusarium sp., Alternaria tenuissima (all from Kazan Federal University Department of Microbiology fungal collection, Kazan, Russia), Alternaria alternata TP 712, Alternaria solani 12RKL15, Doratomyces sp. 14raKKLD and Colletotrichum coccodes 14raKK6 (all from Lomonosov Moscow State University Department of Microbiology fungal collection). The fungi were cultivated in the Czapek medium (NaNO3, 3.0 g; K2HPO4, 1.0 g; MgSO4∙7H2O, 0.5 g; KCl, 0.5 g; FeSO4∙7H2O, 0.01 g; agar, 15 g; distilled water, 900 ml; pH 4.5) [20] . The plates were incubated at 28˚C for 5 - 14 days.
2.3. In Vitro Antagonistic Activity Assay
The interaction studies of rhizobacteria and phytopathogenic fungi were performed using the in vitro dual-culture analysis. Mycelial discs with 8 mm diameter were cut from the target fungi colonies cultured on Czapek plates for seven days and were placed on fresh Czapek plate. Lawns of bacterial individual strains were grown on LB plates for 48 h and were excised from the plate with a sterile scalpel (8 mm). Bacterial blocks were placed at the distance of 3 cm from the fungal discs on the same agar plate. Control plates without bacterial strains were prepared simultaneously. Plates were incubated at 28˚C for 7 - 14 days and examined for the inhibition of fungal growth. To calculate the percent of inhibition we repeated these experiments three times. The growth inhibition of the test fungus was calculated using the following formula:
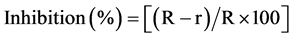 (1)
(1)
where
R―(a control value) represents the radial growth of fungus in control sets.
r―the radial growth of the fungus in sets with bacteria.
2.4. Antagonistic Activity of Extracellular Bacterial Metabolites
Czapek broth was used in studies of antagonistic activity of extracellular bacterial metabolites against the micromycetes. Bacterial cultures were grown in LB broth at 37˚C for 3 days. Cells were removed by centrifugation at 13,000 g for 30 min and then supernatant was filtered through sterile 0.22 microns membrane filter (CAMEO®, GVS, Italy). The resulting supernatant was added to the Czapek broth at the 1 to 4 ratio (10 ml of bacterial culture and 40 ml of Czapek medium). The fungal spores were added at the concentration of 2 × 107 and cultivated aerobically at 28˚C for 6 days on the shaker at 60 rpm (IKA®KS 4000, Germany). Next, fungal cultures were filtered through a filter paper (Whatman No. 1), dried and weighed. Czapek broth without bacterial metabolites and treated in the similar fashion to as described above served as a control. To study temperature stability of metabolites with inhibitory activity bacterial cell-free culture filtrate collected as described above was subjected to autoclave sterilization (30 minutes at 121˚C). The percent of the weight reduction of test fungus was calculated using the following formula:
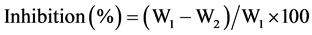 (2)
(2)
where
W1―the weight of the test fungus in control flasks (without bacterial metabolites).
W2―the weight of the fungus in flasks with media containing bacterial metabolites.
2.5. Microscopy of Fungal Mycelia
The morphological changes caused by antagonistic bacteria on the mycelia of the phytopathogenic fungal species (Fusarium sp., A. alternata) after culturing on Czapek plates for 6 - 7 days were directly examined under optical microscope (XSZ-148E, Russia, under 40× magnification).
2.6. Production of Hydrolytic Enzymes and Antifungal Metabolites
Proteolytic activity was determined in well-established plate assay [21] based on the appearance of zones of clearance in the LB-agar containing skim milk (10%). The amylolytic and pectinolytic activities were evaluated on plates containing minimal M9 media [18] supplemented with 0.5% yeast extract and 0.2% soluble starch (v/w) or 0.5% pectin (v/w) to test for amylase and pectinase, respectively. Plates were incubated at 30˚C and after 2 - 3 days were overlaid with 0.1% Lugol’s iodine to identify clear zone formation.
Cellulase activity was assayed on indicator plates containing 0.5% carboxymetylcellulose (CMC) [22] . Plates were incubated at 28˚C for 72 h and then were extensively washed to remove bacteria off the media surface. Next, plates were stained with 1 g∙l−1 Congo Red solution for 30 min. Formation of clear zones around the colonies indicated CMC degradation.
Siderophore production was assessed on chrome azurol S (CAS) agar plates by observing the color change from blue to orange. The plates were incubated at 30˚C for 5 - 7 days [22] .
Ammonia production was tested in colorimetric assay. Addition of 1 ml of Nessler’s reagent to the 72-h-old bacterial culture grown in peptone broth resulted in the development of the yellowish-brown color if ammonia are present in the media [23] .
HCN production of the isolates was detected by the method of Bakker and Schippers [24] . Briefly, HCN production was determined by the color change of the filter paper previously dipped in 2% sodium carbonate/0.05% picric acid solution and placed on the bacterial lawn. Brown color indicated the presence of HCN.
2.7. PCR Amplification of the Genes Responsible for the Synthesis of Antimicrobial Peptides
The genes responsible for the synthesis of antimicrobial metabolites were identified by PCR amplification using specific primers to sequences of genes srfA (surfactin), bacA (bacilysin), fenD (fengycin), bmyB (bacillomycin), ituC (iturin). Partial sequence of ituC (acyl-protein synthetase ItuC) gene from B. subtilis B010 (http://www.ncbi.nlm.nih.gov/nuccore/262182344) with the size of 468 b.p. was used to design primers for ituC gene; B. subtilis QST713 (353 b.p., http://www.ncbi.nlm.nih.gov/nuccore/DQ011330.1) sequence was used for bmyB (peptide synthase BmyB) gene; B. subtilis subsp. spizizenii TU-B-10 (615 b.p., http://www.ncbi.nlm.nih.gov/gene/11241385) sequence was used for bacA (bacilysin biosynthesis protein BacA) gene; B. subtilis subsp. subtilis 168 (10764 b.p., http://www.ncbi.nlm.nih.gov/gene/938306) sequence was used for srfA (surfactin synthase subunit 1 SrfA) gene; B. subtilis (7774 b.p., http://www.ncbi.nlm.nih.gov/nuccore/AJ011849.1) sequence was used for fenD (fengycin synthetase FenD) gene.
Multiple alignments of the genes involved into antimicrobial peptide biosynthesis for sequenced B. subtilis strains present on the NCBI server (http://www.ncbi.nlm.nih.gov/) were performed to design primers to the conservative regions (Table 1).
The annealing temperature of primers was calculated using the program OligoCalc (http://www.bio.bsu.by/molbiol/oligocalc.html). Amplification of these marker genes showed that each gene had one specific band of the expected size.
![]()
Table 1. Primers for antimicrobial peptide biosynthesis genes.
2.8. Lipopeptide Fraction Preparation
B. subtilis strains GM2 and GM5 were grown in 200 ml of LB-broth at 30˚C for 72 h. Bacteria were removed by centrifugation at 12000 g for 30 min at 4˚C followed by filtration of conditioned media through 0.22 µm filters (Millipore). pH was adjusted to 2.5 using 6N HCl. Acidified conditioned media were subjected to centrifugation at 10,000 g for 30 min at 4˚C. Resulting pellets were dissolved in methanol-water (50/50), incubated for 3 h at room temperature and spun down by centrifugation at 10,000 g for 10 min. Methanol-soluble fraction was transferred to a new tube and dried under vacuum at 45˚C for 3 h. Dried samples were dissolved in 50 µl of methanol-water (50/50) and used for HPLC analysis. An equal volume of LB-broth was treated in a similar fashion and used as a negative control.
2.9. HPLC Analysis
Lipopeptides were dissolved in methanol and separated by HPLC using Acclaim® Polar Advantage II (PA2) С18 reverse-phase column (5 µm, 250 × 4.6 mm) and UltiMate 3000 UHPLC system (Thermo Scientific, Dionex, USA) as described by Roy et al. [25] .
2.10. Antifungal Activity of Lipopeptide Fraction
Antifungal activity of HPLC-purified lipopeptide fraction from B. subtilis GM2 and GM5 was determined by disc diffusion assay. Mycelial discs with 8 mm diameter were cut from the target fungi colonies of F. oxysporum and F. solani cultured on Czapek plates for seven days and were placed on fresh Czapek plate. 10 µl of each lipopeptide fraction (see sections Lipopeptide fraction preparation and HPLC analysis) were placed on the sterile filter discs (6.5 mm). Methanol- PBS (50/50) was used as a control. Filter discs were placed 3 cm away from the fungus and plates were incubated at 30˚C for 6 days. Formation of growth inhibition zone around lipopeptide-containing discs indicated the presence of antifungal activity.
2.11. Plant Growth Promotion and Antifungal Activity of B. subtilis GM2 and GM5 on Wheat Seedlings
Wheat seeds were sterilized in 70% ethanol for 30 s, washed three times with sterile water, incubated for 5 min in 2.5% sodium hypochlorite, washed again with sterile water three more times and air dried. Sterilized seeds were placed on sterile filter paper soaked in sterile water and incubated in a Petri dish at room temperature for 24 h. Twenty seedlings were selected for each experimental group. To evaluate plant growth promotion by B. subtilis GM2 and GM5 seedlings were incubated either with water or with 3-days old bacterial suspension (107 CFU/ml) for 1 h, placed on water soaked filter and incubated in sterile Petri dish at 25˚C for 8 days.
To evaluate antifungal activity of B. subtilis GM2 and GM5 8 mm mycelial discs of 7-days-old F. oxysporum (see section In vitro antagonistic activity assay) were placed in 500 ml of sterile water and incubated for 30 minutes at room temperature with shaking (200 rpm) to release spores. Resulting spore suspension was further diluted to 105 CFU/ml. Wheat seedings were placed on the sterile paper filters soaked either in water or in 5 ml of spore suspension and incubated in the Petri dishes for 8 days at room temperature. Seedling survival was expressed in percent of total number of seedlings in each group. Root and shoot dry weights were measured and the average values were calculated and compared to corresponding weights obtained for seedlings from the control group.
3. Results
We isolated 48 bacterial isolates from three independent potato plants rhizosphere characterized by different colony morphology. Twenty five isolates were identified as Gram positive bacteria; the remaining 23 isolates had Gram negative staining. Organism identification by MALDI BioТyper showed that Bacillus species were predominant among Gram positive bacteria: Bacillus subtilis (14 isolates), Bacillus pumilus, Bacillus weihenstephanensis, Bacillus thuringiensis. Other Gram positive bacteria included Lysinibacillus fusiformes, Lysinibacillus sphaericus, Brevibacterium iodinum. Among Gram negative bacteria were identified Acinetobacter calcoaceticus, Citrobacter freundii, Enterobacter amnigenus, Enterobacter cloacae, Enterobacter ludwigii, Myroides odoratus, Providencia alcalifaciens, Serratia rubidaea, Serratia plymuthica, Serratia grimesii, Pseudomonas putida, Alcaligenes faecalis and Brevundimonas diminuta.
Analysis of antagonistic activity of newly isolated strains against phytopatogenic Fusarium sp. showed that 18 Gram positive and 6 Gram negative isolates strongly suppressed the growth of this microscopic fungus. Thirteen out of 18 Gram positive bacteria were identified as B. subtilis.
We selected two bacterial isolates with highest antagonistic activity against Fusarium sp., GM2 and GM5, for further studies. Both strains were gram-posi- tive endospore-forming rods. Oxidase and catalase reactions were positive for both strains.
The identification by Biotyper (MALDI-TOFF analysis) allowed assigning both strains to the Bacillus genus: score value for the strain GM2 was 2.150, and for GM5 ? 2.164. This assignment was further confirmed by 16S rRNA and subunit B of gyrase genes homology. Based on the analysis of 16S rRNA full- gene sequence strain GM2 had the closest similarity to B. subtilis strains JCM 1465, NBRC 13719 and BCRC 10255 (99% of identity to the sequence of the available 1044 bp fragment of 16S rRNA gene). Sequence of the subunit B of gyrase gene was 97% identical to the corresponding gene from B. subtilis subsp. spizizenii TU-B-10 (1011 bp). On the basis of 16S rRNA gene homology strain GM5 was close to the strain B. subtilis 168 (98% for sequence with the size of 1010 bp), and also to strains JCM 1465, NBRC 13719 and BCRC 10255 (98% of homology for 942 bp fragment). By the homology of subunit B of DNA gyrase gene sequence the strain GM5 was also similar to the corresponding gene from B. subtilis strains TO-A JPC, KCTC 1028 and 168 (98% for 978 bp fragment). Thus, we concluded that both isolates are the new strains of B. subtilis.
As shown in Figure 1, the strains B. subtilis GM2 and B. subtilis GM5 demonstrated antagonistic activity against all tested phytopathogenic fungi: F. ave-
![]()
Figure 1. Antagonism of B. subtilis GM2 and B. subtilis GM5 against fungal phytopathogens: (1) Alternaria alternata ТP 712; (2) Alternaria solani 12RKL15; (3) Alternaria tenuissima; (4) Fusarium avenaceum; (5) Fusarium solani; (6) Fusarium oxysporum; (7) Fusarium sp.; (8) Fusarium redolens; (9) Colletotrichum coccodes 14raKK6; (10) Doratomyces sp. 14raKKLD. Growth inhibition of fungal mycelia was examined in dual culture assay on agar plate.
naceum, F. oxysporum, F. redolens, F. solani, Fusarium sp., A. alternata ТP 712, A. solani 12RKL15, A. tenuissima, Doratomyces sp. 14raKKLD and C. coccodes 14raKK6.
Antagonistic activity was evaluated based on the ability to inhibit growth of micromycetes colony compared to the control plates (Table 2). The highest antagonistic activities of both strains were observed against Doratomyces sp. 14raKKLD (GM2―72% and GM5―79%). B. subtilis GM5 had a greater ability to inhibit growth of all listed micromycetes. For example, the strain GM5 inhibited growth of A. alternata ТP 712, F. avenaceum, F. redolens by 72%, 66% and 65%, respectively while GM2 strain inhibited growth of the corresponding fungi by 48%, 54% and 55% respectively.
Next, we tested the ability of bacterial metabolites to inhibit the growth of F. redolens in liquid culture. Growth inhibition of micromycetes in the Czapek broth containing bacillary metabolites (20% of the volume) was calculated by the comparison of amount of micromycetes’ dry biomass with the control cultures grown in media without metabolite addition (Table 3).
To study the effects of heat-treatment on stability of metabolites autoclaved conditioned medium was used. Metabolites of both strains effectively inhibited growth of micromycetes in the liquid medium (by 82% and 88% for GM2 and
![]()
Table 2. Inhibition of mycelial growth by B. subtilis GM2 and B. subtilis GM5 after seven days of co-culture.
![]()
Table 3. Effect of B. subtilis GM2 and B. subtilis GM5 metabolites on the growth of F. redolans.
GM5 strains, respectively). Autoclaving of the conditioned media did not lead to the loss of inhibitory activity (89% of inhibition by the GM5 strain and 80% by GM2 strain). Therefore, the autoclaving of conditioned media did not cause inactivation of metabolites with antifungal activity.
Thus, the strains B. subtilis GM2 and B. subtilis GM5 secrete the metabolites into the growth media, which are capable micromycetes’ growth inhibition both in liquid and solid media.
The B. subtilis GM2 and GM5 strains were analyzed for their ability to produce hydrolytic enzymes (Figure 2) and other antimicrobial metabolites (Table 4).
Both strains had extracellular protease, amylase, pectinase and cellulase activities. Also both strains were able to produce such antimicrobial metabolites as ammonium and HCN. Interestingly, only GM2 strain was capable to secrete siderophores on chrome azurol S (CAS) agar, which was revealed by orange halo formation around colonies after 5 - 7 days of incubation. There was almost no orange halo around colonies of GM5 strain even after 7 days of incubation, which indicates an absence or very low level of siderophore production under growth conditions used in our study (data not shown).
The exometabolites of B. subtilis GM2 and B. subtilis GM5 affected mycelium morphology of Fusarium sр. and A. alternata in different ways. In addition to
![]()
Figure 2. Production of hydrolytic enzymes by B. subtilis GM2 and GM5: I―protease production on skim milk agar, II―amylase production on medium containing 0.2% soluble starch, III―pectinase production on medium containing pectin 0.5%, IV―cellu- lose production on medium containing 0.5% carboxymetyl cellulose.
![]()
Table 4. Production of antifungal metabolites by antagonistic Bacillus subtilis strains.
*+: positive reaction; -: negative reaction.
the effective inhibition of micromycetes’ growth the secreted metabolites of B. subtilis GM5 also cause considerable alterations in mycelium morphology (Figure 3).
There were numerous changes in mycelium morphology noticed in the presence of GM5 strain comparing to the control. Specifically, irregular, distorted, wrinkled or swollen regions were present, as well as there were “swollen” cells that reminded chlamydospores. In the presence of B. subtilis GM2 strain only single round chlamydospores-like cells were detected. No spore formation was noticed in Fusarium sp. grown in co-culture with either of the two B. subtilis strains.
We amplified the genes responsible for biosynthesis of various antimicrobial peptides (AMPs) from the strains’ genomes: ituC (iturin A synthetase C), bmyB (bacillomycin L synthetase B), fenD (fengycin synthetase), srfA (surfactin synthetase subunit 1), bacA (bacilysin biosynthesis protein BacA) by the PCR.
As shown on Figure 4, the presence of specific PCR products as single bands of the expected size indicated the presence of corresponding genes in the genomes of studied strains.
Four out of five genes (ituC, bmyB, bacA and srfA) were detected in the genome of GM2, and only two genes (srfA and fenD) were identified in the genome of strain GM5. Thus, the strain GM2 has a potential ability to synthesize cyclic lipopeptides iturin A, bacillomycin L (belonging to the family of iturin), surfactin, and bacilysin (dipeptide antibiotic). The strain GM5 has a potential to synthesize cyclic antibiotics fengycin and surfactin.
To confirm that lipopeptides play a major role in antifungal activity of B. subtilis isolates we purified lipopeptide fractions from both strains and used them to
![]()
Figure 3. Morphological changes of mycelia of plant pathogenic fungi upon interaction with B. subtilis GM2 and B. subtilis GM5 in dual culture plates. Images 1 and 2 representative of normal mycelia of Fusarium sp. (1) and A. alternata (2). GM2 and GM5: mycelia from co-inoculated with B. subtilis GM2 and B. subtilis GM5. Abnormal conidia germination in presence bacterial isolates. The arrows indicate the formation of swollen chlamydospores-like cells (optical microscopy at 40×).
![]() GM2 GM5
GM2 GM5
Figure 4. Amplification products of AMP genes. Lane M is a molecular weight marker; ituC (iturin A synthetase C), bmyB (bacillomycin L synthetase B), fenD (fengycin synthetase), srfA (surfactin synthetase subunit 1), bacA (bacilysin biosynthesis protein BacA).
inhibit growth of F. solani and F. oxisporum. We found that lipopeptides from B. subtilis GM5 had higher antifungal activity compared to the corresponding fraction from B. subtilis GM2. Furthermore, lipopeptides from both strains inhibited growth of F. solani (Figure 5) more efficiently than the growth of F. oxisporum (data not shown).
Thus, antagonistic activity of B. subtilis GM5 against phytopathogenic fungus F. solani is at least partly due to production of lipopeptides by this strain. Despite the presence of wider repertoire of genes encoding for cyclic lipopeptides in the genome of B. subtilis GM2 strain (Figure 4), HPLC-purified lipopeptide fraction isolated from this strain was less active against F. solani compared to a similar fraction isolated from B. subtilis GM5 strain (Figure 5).
Finally, we used wheat grains to evaluate plant promoting potential of B. subtilis GM2 and GM5 strains and to directly compare their ability to protect wheat against F. oxysporum.
We found that B. subtilis GM2 treatment stimulated both wheat seedling survival and roots and shoots formation. Curiously, B. subtilis GM5 did not show any plant growth promoting properties (Table 5).
As expected, F. oxysporum treatment severely affected both wheat seedling survival and roots and shoots formation. Infected seedlings were covered in F. oxysporum mycelium. Treatment of Fusarium-infected wheat seedlings with B. subtilis GM5 rescued seedling survival to the level of control group and significantly improved roots and shoots formation. In contrast, no protection was observed in the Fusarium-infected group, additionally treated with B. subtilis GM2 (Table 5). Similar results were obtained in the experiments with the tomato plants (data not shown).
![]()
Figure 5. Antifungicidal activity of purified lipopeptide fractions. HPLC-purified lipopeptide fractions were tested for their ability to inhibit growth of F. solani: 1―metha- nol-PBS (50/50); 2, 3―lipopeptides purified from conditioned media of B. subtilis GM2 (2) and GM5 (3), respectively.
![]()
Table 5. Comparative analysis of growth-promoting and antifungal properties of Bacillus subtilis isolates.
*The total number of wheat seedlings used in each experiment was taken for 100%; **The average dry weight of roots and shoots in the control group was taken for 100%.
Thus, only B. subtilis strain GM5 protected plants against F. oxysporum in vivo. Studies to fully evaluate its potential are currently underway.
4. Discussion
The use of beneficial microorganisms in agriculture and other distorted ecosystems can help to protect crops against phytopathogens. It is known that microorganisms associated with plants can promote their growth and development, e.g. due to the growth inhibition of phytopathogenic microorganism [3] [5] . Isolation of new antagonistic strains is necessary to improve biological control methods and restrain plant diseases. Several Bacillus strains have a suppressive effect on growth of certain phytopathogens and could be used as biocontrol agents [5] [8] [7] . Some members of Bacillus genus have high potential for usage as biological control agents against various plants diseases (Kumar et al. 2012). For instance, B. subtilis strains have demonstrated antagonistic activity against Fusarium sp. [10] [26] .
In this study, two strains with high antagonistic activity against micromycetes were isolated from potato’s rhizosphere.
MALDI-TOF analysis assigned isolated strains GM2 and GM5 to Bacillus genus and predicted that they belong to B. subtilis species. An accurate species assignment of isolates using this technique could be done when the Score values are in the range 2.3 - 3.0. Further studies based on 16S rRNA and gyrase subunit B genes homology confirmed that isolated bacteria represent new strains of B. subtilis.
It is known that B. subtilis species are heterogeneous both phenotypically and genotypically [27] . High genetic heterogeneity of different Bacillus species [5] [28] particularly B. subtilis allows to suggest that search and identification of new strains from different sources may expand the number of practically important strains and to improve our understanding of mechanisms involved in antagonistic interactions.
Comparative analysis of antagonistic activity of isolated B. subtilis isolates showed that strain GM5 inhibits the growth of various micromycetes more effectively than the strain GM2 especially on solid media. The inhibitory activity of extracellular metabolites was not affected by sterilization of Bacillus-conditioned media by autoclaving, which suggests that the main antifungal activity is associated with thermo-stable metabolites. It is known that Bacillus ssp. protect plants against phytopathogenic bacteria and micromycetes through a number of mechanisms, in particular through the synthesis of different cyclic lipopeptides with inhibitory activity against phytopathogens [29] . The genes responsible for biosynthesis of antimicrobial peptides were identified in different species of Bacillus, and mostly in B. subtilis and B. amyloliquefaciens strains [29] . The analysis of large number of Bacillus spp. isolates (184) associated with plants showed that the significant number of strains possessed at least 2 to 4 genes encoding for antimicrobial peptides. Majority of strains carry srfA, bacA, bmyB, fenD genes [30] . Thus, pre-screening for the presence of these genes could be useful for selection of potentially promising strains for biocontrol of phytopathogens.
We have shown that two newly isolated strains of B. subtilis differ in the repertoire of genes encoding for antimicrobial peptides. Cyclic lipopeptides (CLP) produced by B. subtilis strains have been shown to protect host plants from a numbers of pathogens. CLP produced by B. subtilis can be grouped into three families: surfactins, fengycins, and iturins, each of which has shown different antimicrobial characteristics [9] . PCR-screening revealed that the strain GM2 contains four genes responsible for synthesis of cyclic lipopeptides like iturin A, bacillomycin L (belongs to the iturin family) and surfactin and also dipeptide antibiotic bacilysin. The strain GM5 has two genes necessary for synthesis of cyclic lipopeptides surfactin and fengycin. Higher antagonistic activity of GM5 correlated with the presence of fengycin gene that was not identified in GM2 strain. Moreover, purified lipopeptide fraction from GM5 strain showed significantly higher ability to inhibit fungal growth compare to a similar fraction purified from GM2 strain. Fengycins are cyclic lipodecapeptides which specifically inhibit growth of filamentous fungi [31] . Fengycin draws a lot of attention in the recent years because of its lower hemolytic activity compared to other B. subtilis lipopeptides and its strong antifungal capability, specifically against filamentous fungi [32] . Fengycin is known to affect the biological membranes of fungi, causing membrane pore formation, however, the precise mechanism of fengycin’s action and the intrinsic biochemical determinants implicated in its antifungal activity have not been fully elucidated [31] . Simultaneous production of fengycins and surfactins could be important for the efficiency of F. graminearum control by strain B. subtilis SG6 [26] .
There were significant changes in the mycelium morphology caused by bacteria in comparison to the control: irregular, distorted, shrunken or swollen areas appeared; many “swollen” chlamydospore-like cells were seen. The appearance of such anomalous formations in mycelium in the presence of metabolites of bacterial antagonists was frequently noticed by others [33] [34] . Antifungal action of Bacillus metabolites may be due to disruption of fungal cell wall and inhibition of normal conidia development. It is known that chlamydospores with thick walls are developed by the modification of hyphal and conidial cells under unfavorable environmental conditions, such as at low temperature, and these resting bodies act as primary inocula in subsequent soil-borne infections, whereas the microconidia and macroconidia formed by F. oxysporum are important in secondary infection [35] [36] . Minerdi et al. [37] reported that microbial symbionts silence the virulence of F. oxysporum and that changes in cell morphogenesis by F. oxysporum underlie this suppression.
Various bacterial features were examined to identify other secondary metabolites and additional factors which may correlate the differences in antagonistic activity of studied strains. It is known that extracellular hydrolases make important contributions to the antimicrobial potential of antagonists. It was shown that synthesis of extracellular hydrolases correlates with antifungal potential because some hydrolytic enzymes are involved in the degradation of phytopathogenic micromycetes’ cell wall [10] [38] . It was also suggested that pectinases and amylases contribute to the colonization of roots by bacteria and therefore may play an important role in plant growth stimulation [28] . We have shown that both strains secrete proteinases, amylases, pectinases and cellulases to the conditioned media.
We have found that out of two B. subtilis strains only one of them, GM2, produced significant amount of siderophores as indicated by formation of orange halos around the colonies on the CAS agar medium. It was suggested that the synthesis of siderophores is associated with growth-promoting and antagonistic activity against phytopathogens [3] . In the soil, ability to produce siderophores plays a central role in the capacity of different bacteria to promote plant development due to increasing the iron absorption by plants and defense of plants from toxic metals such as nickel and cadmium [39] [40] . The antagonism against pathogenic bacteria and fungi is based on the iron binding by siderophore-producing bacteria that limits iron availability for other microorganisms and primarily for phytopathogenic micromycetes [41] . Both strains produce cyanides and ammonium that could contribute to their antifungal activity. It is known that cyanogens produced by Bacillus can inhibit fungal growth [4] .
Nevertheless, our findings showed that the ability to secrete highly active proteinases and to produce siderophores does not correlate directly with the antifungal potential of the Bacillus strains. While siderophore-producing strain GM2 expressed plant growth promoting properties, only B. subtilis GM5 strain was able to protect wheat seedlings and tomato plants against F. oxysporum, an important pathogen of wheat [42] [43] and tomato [44] .
5. Conclusion
We have isolated two new strains of B. subtilis that differ in their antagonistic activity against a number of phytopathogenic fungi. Only B. subtilis GM5 strain was able to protect wheat from phytopathogenic fungus Fusarium oxysporum in vivo. The antifungal activity of GM5 strain was attributed with secretion of antimicrobial peptides. We suggest that a high antifungal activity might be associated with the presence of fenD gene identified only in GM5 strain. Thus, B. subtilis GM5 can find application in agriculture as bioinoculant for agriculturally important plants.
Acknowledgements
This work was performed in accordance with the Russian Government Program of Competitive Growth of the Kazan Federal University. This work was supported by the subsidy allocated to the Kazan Federal University for the state assignment in the sphere of scientific activities (Project No. 14-83 0211/02.11. 10083.001). The experiments on the characterization of extracellular hydrolytic activity of bacilli were carried out on means of Russian Science Foundation (Project No. 16-16-04062).
Competing Interests
The authors declare that there are no competing interests regarding the publication of this paper.