Identifying Semantic in High-Dimensional Web Data Using Latent Semantic Manifold ()
1. Introduction
In the traditional approach to data gathering, we collect data on a few well-chosen variables, and then manually perform various tasks, such as finding relevant information, analyzing them, making decisions, and so on [1] . However, in this high-tech era, the high volumes of data are generated with high velocity from a variety of re- sources (also known as 3 V―Volume, Velocity, and Variety) [2] [3] . The modern Information and Communica- tion Technology (ICT) infrastructure, the advent of cloud computing, the cheaper availability of storage device, and the low cost computing power have made people capable of recording and storing an enormous amount of data [4] . As a result, gigantic repositories that include data, texts, and media have rapidly grown during recent years [5] - [9] . Nowadays, we create as much information in every two days as we have done since the dawn of civilization [10] [11] . Several huge repositories are freely available for the public use on the World Wide Web causing another problem―the relevant information is buried in the irrelevant ones.
To combat the problem to lose the relevant information in the overwhelming amount of data, a number of search engines have proliferated recently, which can aid users in searching contents which are relevant to them [8] . As the web pages are heterogeneous and consist of varying quality, they put limitations on search technologies [12] [13] . Moreover, the relationships among the words (polysemy, synonymy, and homophony) and sentences (paraphrase, entailment, and contradiction), and ambiguities (lexical and structural) diminish the search engines’ power [14] [15] . Hence, the search engines often return inconsistent, uninteresting, and unorganized results [9] [12] . Web users have to devote substantial time and effort to differentiate meaningful items from the results returned by the search engines [9] [16] [17] . In order to facilitate and enhance relevant information access to the web users, it is essential for search engines to deal with ambiguities and imprecision [18] [19] . The need to enhance the search engines’ capabilities has been felt such that the search engines can not only generate results of the web users’ queried terms, but also can filter and organize meaningful items from the irrelevant ones [20] - [22] .
Many effective search engines, such as MedEvi, EBIMed, MEDIE, PubNet, GoPubMed, Argo, and Vivisimo, have provided capabilities to fit search results to the users’ intent. These search engines can discover latent semantic (relationships between a set of documents and the terms they contain) in the search engine generated documents and classify these documents into homogeneous semantic clusters [23] - [33] . In these search engines, each semantic cluster is considered as a topic, which indicates a summary of the generated documents. Later, the users can explore the topics that are relevant to their intent. For example, upon using these search engines, a query term, APC (Adenomatous Polyposis Coli), can yield abstracts of the relevant PubMed articles. In this case, the generated results will consist of not only abstracts about Adenomatous Polyposis Coli, but also others such as Antigen Presenting Cells (APC), Anaphase Promoting Complex (APC), and Activated Protein C (APC). The users need to find articles which are relevant to their intent (here Adenomatous Polyposis Coli) after going through the abstracts generated from the search. In summary, rather than providing huge number of web links related to the queried terms, search engines need to generate results relevant to users’ intent.
In the past, many algorithms/techniques have been deployed to develop semantic search engines as described in the previous paragraph [25] . For instance, deterministic search techniques have provided metadata-enhanced search facility, where a user pre-selects different facets to generate more relevant search results [18] [19] . However, scaling metadata-enhanced search facility to the web is difficult and requires many experts to define controlled-vocabulary in order to create unique labels for the concepts having the same terminology [34] [35] . Luhn pointed out that the frequency of terms and their relative positions within a sentence in a document can be used to compute a relative measure of significance, first for the individual words and then for the sentences [36] . Word usage in a document collection tends to follow Zipf’s distribution, in which a few words are used very frequently, but the vast majority only rarely [37] . Therefore, Salton and McGill addressed the 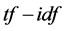 scheme, which is a measure of each basic element (term) in a document collection to reveal the significance of elements within the collection [38] . For each document in the collection, the
scheme, which is a measure of each basic element (term) in a document collection to reveal the significance of elements within the collection [38] . For each document in the collection, the 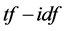 value of each term is determined by the term frequency, that is, the number of occurrences of each term in the document but is offset by the frequency of the word in the corpus, which helps to adjust for the fact that some words appear more frequently in general. We may view each document as a vector with one component corresponding to each term together with a weight for each component. Thus, the
value of each term is determined by the term frequency, that is, the number of occurrences of each term in the document but is offset by the frequency of the word in the corpus, which helps to adjust for the fact that some words appear more frequently in general. We may view each document as a vector with one component corresponding to each term together with a weight for each component. Thus, the  scheme can reduce documents of arbitrary length to fixed- length lists of numbers. The
scheme can reduce documents of arbitrary length to fixed- length lists of numbers. The 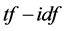 weighting schemes are often used by search engines as a central tool in scoring and ranking a document’s relevance given a user query. In addition,
weighting schemes are often used by search engines as a central tool in scoring and ranking a document’s relevance given a user query. In addition, 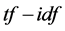 can be successfully used for stop-words filtering in various subject fields including text summarization and classification. No doubt, the revolutionary change was realized in the information retrieval field with the introduction of
can be successfully used for stop-words filtering in various subject fields including text summarization and classification. No doubt, the revolutionary change was realized in the information retrieval field with the introduction of  scheme and its variants. However, in the
scheme and its variants. However, in the 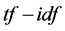 scheme, the document collection is presented as a document-by-term matrix, which is usually enormously high-dimensional and sparse [38] - [40] . Often, for a single document, there are more than thousands of terms in the matrix, and most of the entries are zero. The
scheme, the document collection is presented as a document-by-term matrix, which is usually enormously high-dimensional and sparse [38] - [40] . Often, for a single document, there are more than thousands of terms in the matrix, and most of the entries are zero. The  scheme can bring down some terms; yet, it provides a relatively small amount of reduction, which is not enough to reveal the statistical measures within a document or between documents.
scheme can bring down some terms; yet, it provides a relatively small amount of reduction, which is not enough to reveal the statistical measures within a document or between documents.
In the last decades, some other dimension reduction techniques, such as Latent Semantic Indexing, Probabilistic Latent Semantic Indexing, and Latent Dirichlet Allocation models have been proposed to overcome the shortcomings of earlier search engines. But, all these are based on bag-of-words models. The bag-of-words models follow Aldous and de Finetti theorem of exchangeability where the order of terms in a document or order of documents in a corpus can be neglected [41] - [43] . As the spatial information conveyed by the terms in the document or documents in the corpus was highly neglected in these approaches, we found a statistical issue attached with these bags-of-words models [42] - [45] . In the probability theory, the random variables (here referred as terms) 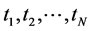 are said to be exchangeable if the joint distribution
are said to be exchangeable if the joint distribution 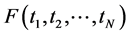 is invariant under permutation of its arguments, so that
is invariant under permutation of its arguments, so that 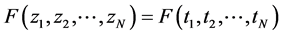 whenever
whenever 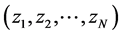 is a permutation of
is a permutation of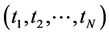 . However, these terms are exchangeable and the relationship between them can be established if the terms are located in proximity. For instance, we have a document describing products, such as laptops, mobile phones, and notepads. The appearance of the word “apple” can be associated with a company if it appears in proximity to words laptop, mobile phone, and notepad. However, in case, the word “apple” appears after several words or pages in the document, the relationship between “laptop, mobile phone or notepad” and “apple” weakens. Therefore, the criteria-the order of terms in a document can be neglected-should be modified to order of terms in a relationship of a document can be neglected. Likewise, the order of documents in a corpus can be neglected should be modified to the ordering documents in relationships of a corpus can be neglected. For instance, a search term “network” would yield different topics if it occurs nearby to a term, such as computer, traffic, artificial neural, or biological neural; and hence, the order of in-relationship terms might be neglected [46] .
. However, these terms are exchangeable and the relationship between them can be established if the terms are located in proximity. For instance, we have a document describing products, such as laptops, mobile phones, and notepads. The appearance of the word “apple” can be associated with a company if it appears in proximity to words laptop, mobile phone, and notepad. However, in case, the word “apple” appears after several words or pages in the document, the relationship between “laptop, mobile phone or notepad” and “apple” weakens. Therefore, the criteria-the order of terms in a document can be neglected-should be modified to order of terms in a relationship of a document can be neglected. Likewise, the order of documents in a corpus can be neglected should be modified to the ordering documents in relationships of a corpus can be neglected. For instance, a search term “network” would yield different topics if it occurs nearby to a term, such as computer, traffic, artificial neural, or biological neural; and hence, the order of in-relationship terms might be neglected [46] .
As we can see from the literature review and our arguments that there is a need to enhance search engines’ capabilities to reveal latent semantics in high-dimensional web data while preserving the relationship and order of term(s) or document(s). We proposed a novel algorithm called Latent Semantic Manifold (LSM), which identifies homogeneous groups in web data while preserving the spatial information about terms in a document or documents in the corpus. This paper aims to explain the Latent Semantic Manifold algorithm (from now on, LSM algorithm), its deployment, and performance evaluation.
2. Methods
This study consists of three key components: proposing and describing the LSM algorithm, its deployment, and evaluation. They are described in the following subsections.
2.1. Algorithm
The proposed LSM algorithm is based upon the concepts of probability and topology, which identifies the latentsemantic in data. Figure 1 and Table 1 provide the high-level view of the algorithm. The concepts deployed in the LSM algorithm are explained in the following four steps.
Step 1 (Identifying relevant fragment from the user query generated documents): A user can enter a query using a search engine, which generates a set of documents. The relevant fragments (paragraphs in the LSM) are identified from the generated documents. The identification of the fragments is handled by the “document preprocesssor” of the search engine, which typically normalizes the document stream to a predefined format, breaks the document stream into desired retrievable unit, and isolates and metatags subdocument pieces.
Step 2 (Recognizing named-entity and constructing heterogeneous manifold): It is crucial to extract significant “terms” from the fragments (identified in Step 1) to construct heterogeneous manifolds. Notably, we can extract various types of terms with a large number of training documents. However, extracting different types of terms and calculating their marginal and conditional probabilities is highly computation-intensive [47] - [51] . Therefore, we stick to identifying nouns (words or phrases) or named-entities in the LSM framework. Hidden Markov Mod- els (HMMs) are often used for part-of-speech tagging and sequential labeling [52] [53] . Yet, in the last decade, discriminative linear chain Conditional Random Field (CRF) models have been used for tagging and sequential labeling of features in the corpus because of its advantages over the HMMs [54] - [56] . The primary advantage of CRFs over HMMs is their conditional nature. A CRF is a simple framework for labeling and segmenting data that
![]()
Figure 1. Illustration of LSM algorithm.
models a conditional distribution P(z|x) by selecting the label sequence z, a named category, to label a novel observation sequence x with an associated undirected graph structure that obeys the Markov property. When
![]()
Table 1. LSM algorithm that construct semantic manifold.
conditioned on the observations that are given in a particular observation sequence, the CRF defines a single log-linear distribution over the labeled sequence. The CRF model does not need explicitly to present the dependencies of input variables x affording the use of rich and global features of the input, thus allows relaxation of the strong independence assumptions required by HMMs in order to ensure tractable inference. The relationships among these named-entities construct a complex structure called a heterogeneous manifold.
The named-entities are indicated with their marginal probabilities, and the correlations among named-entities are indicated with their conditional probabilities. For example, the jaguar is considered as a named-entity, and it is assigned to the animal or vehicle type depending on the overall context of the fragment. The named-entities are indicated with their marginal probabilities, and the correlations among the named-entities are indicated with their conditional probabilities. As illustrated in Figure 2, Jaguar is a named-entity with three possible types-animal, vehicle, and instrument. It has marginal probabilities, such as Panimal (Jaguar), Pvehicle (Jaguar), and Pinstrument (Jaguar). Likewise, it has conditional probabilities, such as P (Jaguar, Car|Vehicle), P (Jaguar, Motorcycle|Vehicle).
Step 3 (Decomposing a heterogeneous manifold into homogeneous manifolds): As mentioned in Step 2, the he- terogeneous manifold consists of a complex structure of named-entities, including estimates of marginal and con- ditional probabilities. A collection of fragment vectors lies on the heterogeneous manifolds, which contains some local spaces resembling Euclidean spaces of a fixed number of dimensions. Every point of the n-dimensional he- terogeneous manifold has a neighborhood homeomorphic to the n-dimensional Euclidean space Rn. In addition, all the points in the local spaces are strongly connected. As the heterogeneous manifold is overly complex, and the semantic is latent in local spaces; thus, instead of retaining just one heterogeneous manifold, we break it into a collection of homogeneous manifolds. The topological and geometrical concepts can be used to represent the la- tent semantics of a heterogeneous manifold as a collection of homogeneous manifolds. A Graph-based Tree-width Decomposition algorithm is used to decompose a heterogeneous manifold into a collection of homogeneous ma- nifolds [57] . As shown in Figure 3, assuming Jaguar as the heterogeneous manifold, we can decompose it into three homogeneous manifolds bounded by dotted lines in three different colors. In the Graph-based Tree-width Decomposition algorithm, we start selecting a random fixed dimension local manifold to be a separator as shown in Figure 4 [58] . Afterward, the local manifold is decomposed into two local manifolds that are not adjacent. This decomposition is recursive until no further decomposition is possible. We can express the above concept formally,
![]()
Figure 2. An example to demonstrate named-entities, its types, and associated marginal and conditional probabilities.
![]()
Figure 3. An example to demonstrate Graph-based Tree-width Decomposition.
![]()
Figure 4. An example to demonstrate the concept of separator under Graph-based Tree-width decomposition.
let a heterogeneous manifold Mi for fragmenti be the set of homogeneous manifolds, such that
![]() . The semantics generated from fragment homogeneous manifolds
. The semantics generated from fragment homogeneous manifolds
are independent. In addition, a semantic topic set ![]() . of the returned documents is associated
. of the returned documents is associated
with semantic mapping ![]() with a probability
with a probability![]() , and quantity
, and quantity![]() . The
. The
probabilities indicate the number of documents that are relevant to a homogeneous manifold and match the user’s intent. To induce homogeneous manifolds, it is crucial to extract significant terms from fragments. In addition, we should demonstrate the relevance of each fragment to the homogeneous manifold. The users can refer only homogeneous manifold associated fragments, which they want.
Step 4 (Exploring the homogeneous manifolds): The relevant fragments cluster around their related homogeneous manifolds. For instance, a user query for the term APC, the fragments have aggregated into a collection of homogeneous manifolds as shown in Figure 5. Each fragment is assigned to a particular homogeneous manifold.
2.2. Deployment of the LSM Algorithm
The LSM algorithm was deployed to develop a search tool. A team of three researchers including an expert in the Java programming language developed the tool using the Eclipse Software Development Kit. The LSM tool was used for two years at two places in Taiwan: 1) Taipei Medical University Library, Taipei; and 2) Biomedical Engineering Laboratory, Institute of Biomedical Engineering, National Taiwan University, Taipei. The members of the library and lab used the LSM tool to perform semantic searches in the PubMed database.
2.3. Performance Evaluation of the LSM Algorithm
Data sets: Two data sets, Reuters-21578-Distribution-1 and OHSUMED, were used to evaluate the performance
![]()
Figure 5. An example to demonstrate heterogeneous manifold, homogeneous manifolds and documents associated with homogeneous manifolds.
of the LSM algorithm. The Reuters-21578-Distribution-1 is a standard benchmark for the text categorization, which consists of Newswire articles classified into 135 topics [59] . In our tests, the documents with multiple topics (category labels) and single topic were separated. The topics that had less than five documents were removed. Table 2 shows the summary of the Reuters-21578-Distribution-1 collection. OHSUMED is clinically oriented a Medline collection consisting of 348,566 references. It covers all the references from 270 medical journals belonging to 23 disease categories over a five-year period (1987-1991) [60] .
Evaluation criteria: Effectiveness and efficiency were measured as an experimental evaluation of the LSM algorithm. Effectiveness is defined as the ability to identify the right cluster (collection of documents). As shown in Table 3, the generated clusters were verified by human experts to measure the effectiveness. The three measures of the effectiveness (Precision, Recall, and![]() ) were calculated using the contingency in Table 3. Precision and Recall are respectively defined as follows:
) were calculated using the contingency in Table 3. Precision and Recall are respectively defined as follows:
![]()
![]()
Moreover, ![]() measure, which combines Precision and Recall, is defined as follows:
measure, which combines Precision and Recall, is defined as follows:
![]()
![]() measure is used in this paper, which is obtained assigning
measure is used in this paper, which is obtained assigning ![]() to be 1, which means that precision and recall have equal weight in evaluating the performance. In case, many categories are generated and compared, the overall precision and recall are calculated as the average of all precisions and recalls belonging to various categories. F1 is calculated as the mean of all results, which is a macro-average of the categories.
to be 1, which means that precision and recall have equal weight in evaluating the performance. In case, many categories are generated and compared, the overall precision and recall are calculated as the average of all precisions and recalls belonging to various categories. F1 is calculated as the mean of all results, which is a macro-average of the categories.
In addition, two other evaluation metrics, Normalized Mutual Information (NMI) and overall F-measure, were also used [61] - [63] . Given the two sets of topics C and Cl, let C denote the topic set defined by experts, and Cl denotes the topic set generated by a clustering method, and both derived from the same corpora X. Let N(X) denote the number of total documents; N(z, X) denotes the number of documents in topic z; and N(z, z', X) denotes the number of documents both in topic z and topic z', for any topics in C. The Normalized Mutual Information metric MI(C, C') is defined as:
![]()
where ![]()
![]()
Table 2. Statistics of reuters-21,578 corpora.
![]()
Table 3. Contingency table for category (ci, where i = natural number)a.
aTP: True Positive; FP: False Positive; FN: False Negative; TN: True Negative.
The Normalized Mutual Information metric MI(C, C') will return a value between zero and![]() , where H(C) and H(C') define the entropies of C and C' respectively. A higher
, where H(C) and H(C') define the entropies of C and C' respectively. A higher ![]() value means that two topics are almost identical, whereas a lower value indicates the independence of topics. Therefore, the Normalized Mutual Information metric
value means that two topics are almost identical, whereas a lower value indicates the independence of topics. Therefore, the Normalized Mutual Information metric ![]() is
is
![]()
Let ![]() be an F-measure for each cluster
be an F-measure for each cluster ![]() defined above. The overall F-measure can be defined as
defined above. The overall F-measure can be defined as
![]()
where F(z, z') calculates the F-measure between z and z'.
Efficiency is the clustering time for a search query with a fixed number of features for each clustering scheme, where features set is fixed.
Experiments: The experiments were conducted using Reuters-21578-Distribution-1 and OHSUMED data sets. The clusters ranging from two to ten topics were randomly selected to evaluate the LSM with other clustering methods. For each clustering method, each test run was conducted on a selected topic, and Normalized Mutual Information of the topic and its corresponding cluster was calculated. After conducting fifty test runs on a fixed number of k’s topics, where![]() , the final performance scores were obtained by averaging mutual in- formation measures from these 50 test runs [61] . The t-test assessed whether homogeneous clusters generated by the two methods (LSM vs. other methods) were statistically different from each other as shown in Table 4 and Figure 6 in the result section. We also calculated the overall F-measure in combination of arbitrary k clusters by uniquely assigning to topics from the Reuters-21578-Distribution-1 data set where k was 3, 15, 30, and 60 [64] . Fifty test-runs were also performed using these LSM results to compare Frequent Itemset-based Hierarchical Clustering (FIHC) and bisecting k-means as shown Table 5 and Figure 7 in the Result section [64] [65] .
, the final performance scores were obtained by averaging mutual in- formation measures from these 50 test runs [61] . The t-test assessed whether homogeneous clusters generated by the two methods (LSM vs. other methods) were statistically different from each other as shown in Table 4 and Figure 6 in the result section. We also calculated the overall F-measure in combination of arbitrary k clusters by uniquely assigning to topics from the Reuters-21578-Distribution-1 data set where k was 3, 15, 30, and 60 [64] . Fifty test-runs were also performed using these LSM results to compare Frequent Itemset-based Hierarchical Clustering (FIHC) and bisecting k-means as shown Table 5 and Figure 7 in the Result section [64] [65] .
![]()
Table 4. Normalized Mutual Information comparison of LSM algorithm with other sixteen methods using Reuters-21578- Distribution-1 datasetb.
bLSM: Latent semantic manifold; CCF-k: clique community finding algorithm; GMM: Gaussian mixture model; NB: Naive Bayes clustering; GMM + DFM: Gaussian mixture model followed by the iterative cluster refinement method; KM: Traditional k-means; KM-NCL Traditional k-means and spectral clustering algorithm based on normalized cut criterion; SKM: Spherical k-means; SKM-NCW: Normalized-cut weighted form; BP-NCW: Spectral clustering based bipartite normalized cut; AA: Average association criterion; NC: Normalized cut criterion; RC: Spectral clustering based on ratio cut criterion; NMF: Non-negative matrix factorization; NMF-NCW: Nonnegative Matrix Factorization-based clustering; CF: Concept factorization; CF-NCW: Clustering by concept factorization.
![]()
Table 5. Precision, recall, overall F-measure, and Normalized Mutual Information (NMI) of Latent Semantic Manifold on Reuters-21578-Distribution-1 dataset.
![]()
Figure 6. Mutual information values of 2 to 10 clusters built by LSM algorithm and other sixteen methods using Reuters-21578-Distribution-1 datasetsc. c. LSM: Latent semantic manifold; GMM-Gaussian mixture model; NB: Naive Bayes clustering; GMM + DFM: Gaussian mixture model followed by the iterative cluster refinement method; KM: Traditional k-means; KM-NC: Traditional k-means and spectral clustering algorithm based on normalized cut criterion; SKM: Spherical k-means; SKM-NCW: Normalized-cut weighted form; BP-NCW: Spectral clustering based bipartite normalized cut; AA: Average association criterion; NC: Normalized cut criterion; RC: Spectral clustering based on ratio cut criterion NMF: Non-negative matrix factorization; NMF: NCW-Nonnegative Matrix Factorization-based clustering; CF: Concept factorization; CF-NCW: Clustering by concept factorization; CCF: k-clique community finding algorithm.
The average precision, recall, overall F-measure, and Normalized Mutual Information of LSM, LST, PLSI, PLSI + AdaBoost, LDA, and CCF were evaluated using the Reuters-21578-Distribution-1 data set; and LSM, LST, and CCF were evaluated on an OHSUMED data set, as shown in Table 6, in the Result section [44] [66] - [69] . Besides the effectiveness, the efficiency tests of LSM, LST, and CCF were performed as shown in Figure 8 in the Result section.
3. Results
Normalized Mutual Information comparison of the LSM algorithm with the other sixteen methods using Reuters-21578-Distribution-1 data setis shown in Table 4 and Figure 6 [61] [69] - [71] . The four metrics (precision, recall, overall F-measure, and Normalized Mutual Information) of LSM that used Reuters-21578-Distribution-1 data set for different k are listed in Table 5. In addition, the overall F-measure is compared with FIHC and bisecting k-means as shown in Figure 7. The average precision, recall, overall F-measure, and Normalized Mutual Information of 1) LSM, LST, PLSI, LDA, and CCF, which used Reuters-21578-Distribution-1 data set; 2) LSM, LST and CCF, which used OHSUMED data set are shown in Table 6. The efficiency tests results of the three methods, LSM, LST, and CCF, are shown in Figure 8.
![]()
Figure 7. Overall F-measure of three methods, LSM, FIHC, and bisecting k-means, on Reuters-21578-Distribution-1 data set, where k (x-axis) is 3, 15, 30, 60 clusters.
![]()
Figure 8. Efficiency of three clustering methods, wherein x-axis is the number of features and y-axis is run time in milliseconds (LSM: Latent semantic manifold; LST: Latent Semantic Topology; CCF: k-Clique Community Finding).
![]()
Table 6. The average precision, recall, overall F-measure, and Normalized Mutual Information (NMI) of LSM, LST, PLSI, PLSI + AdaBoost, LDA, and CCF on Reuters-21578-Distribution-1 dataset; and LSM, LST and CCF on OHSUMEDd.
dLSM: Latent semantic manifold; LST: Latent semantic topology; PLSI: Probabilistic latent semantic indexing; PLSI + AdaBoost: Probabilistic latent semantic indexing + additive boosting methods; LDA: Latent Dirichlet allocation; CCF: k-clique community finding algorithm.
4. Discussion
Our findings suggest that the LSM algorithm, which can discover the latent semantics in high-dimensional web data, might play an instrumental role in enhancing the search engine functionality. LSM carries out searches based on both keywords and meaning, which can assist researchers to perform semantic searches on databases. For example, a researcher can search APC with Adenomatous Polyposis Coli as his or her intended meaning in the PubMed database (the output of a user queried term APC is shown in Figure 9).
APC can also have other meanings, such as Antigen-Presenting Cells, Anaphase Promoting Complex, or Activated Protein C. Suppose, in a homogeneous manifold, we find APC, Colorectal Cancer, and gene-related documents are assembled, the homogeneous manifoldwould point out the meaning of APC as Adenomatous Polyposis Gene. Similarly, suppose APC, Major Histocompatibility Complex, and T-cells-related documents are assembled, it would indicate the meaning of APC as Antigen Presenting Cells. Figure 9 shows that documents returned from the queried term APC can automatically associate to various homogeneous manifolds (semantic topics). In addition, the researcher can obtain a different vantage point based on the underlying data. For example, a search for the medical term NOD2 that was performed within the PubMed database retrieved almost 300 abstracts of published or in-press articles (Figure 10 shows latent semantic topics as a clustering result).
According to the result, inflammatory bowel disease and its type (Crohn’s disease and ulcerative colitis) are associated with gene NOD2. The term NOD2 was found to be evenly spread over these three topics-inflamma- tory bowel disease, Crohn’s disease, and ulcerative colitis. Some evolving topics, such as the bacterial component were also discovered. However, the result was different when we searched NOD2 on Genia Corpus (Figure 11) which supports the argument the researcher can obtain a different meaningful vantage point based on the underlying data, using the “same” LSM algorithm [72] .
We can see that results (Figure 10 and Figure 11) are meaningfully structured with a possibility of semantic navigation in both databases. This indicates that the generalization capability of the LSM algorithm. We used concepts of topology in designing LSM algorithm. LSM has shown much better performance than the other sixteen clustering methods, especially when the number of clusters gets larger (Table 4 and Table 5, and Figure 6 and Figure 7). In general, we found that LSM could produce more accurate results than others could. We used paired t-test to assess the clustering results of the same topics by any two methods-LSM, LST, and CCF. The results of LSM were significantly better than the results of LST where we used 63 clusters in the experiments
![]()
Figure 10. Clustering result of the query term, NOD2, retrieved from PubMed.
![]()
Figure 11. Clustering result of the query term, NOD2, retrieved from Genia Corpus.
(p-value < 0.05) (Table 6). Similarly, with a p-value less than 0.05, the results of LSM were significantly better than the results of the CCF in 48 randomly selected clusters out of 72 (Table 6). The efficiency evaluation of three methods, LSM, LST, and CCF, demonstrated that LSM performed better than the others did. In the case of LSM, the time needed to build a latent semantic manifold does not increase significantly when the data became larger (Figure 8).
Limitation and future studies: This study has a few limitations that open up the scope of future studies. First, to identify and discriminate the correct topics in the collection of documents, a combination of features and their co-occurring relationships serve as clues, and probabilities display their significance. All features in documents comprise a topological probabilistic manifold, associate to probabilistic measures, and denote the underlying structure. This complex structure is decomposed into inseparable components at various levels (in various levels of skeletons) so that each component corresponds to topics in the collection of documents. This process is a computation-intensive and time-consuming, which strongly depend on features and their identifications (named- entities). Second, some terms with similar meanings, such as anticipate, believe, estimate, expect, intend, and project, were separated into independent topics. Likewise, some terms were repeatedly specified in many topics. These issues might be addressed by utilizing thesauri and some other adaptive methods [73] . Third, some tools, such as MedEvi, EBIMed, MEDIE, PubNet, GoPubMed, Argo, and Vivisimo, also perform a latent semantic search in high dimensional web data [23] - [33] . However, in this study, we did not compare LSM algorithm based tool with others. Some further study is needed to compare the LSM algorithm based tool with already existing tools to find a space of synergy. Fourth, in this study, the evaluation was carried out mainly by comparing with other latent semantic indexing (LSI) algorithms. However, many alternative approaches for searching, clustering, and categorization exist. Further study is needed to compare this approach with alternatives. Fifth, there are some already existing knowledge bases or resources in the biomedical domain, such as MeSH (Medical Subject Headings) [74] [75] . Some studies are needed to verify whether LSM algorithm based tool might be adapted to the existing knowledge bases or resources.
5. Conclusion
We found that the LSM algorithm can discover the latent semantics in high-dimensional web data and can organize them into several semantic topics. This algorithm can be used to enhance the functionality of currently available search engines.
Acknowledgements
The National Science Foundation (NSC 98-2221-E-038-012) supported this work.