Three New Compounds Derived from Nitrofurantoin: X-Ray Structures and Hirshfeld Surface Analyses ()
1. Introduction
Noncovalent weak intermolecular interactions have gained much attention due to their unquestionable role in various chemical, physical, and biological processes [1] -[6] . Among these interactions, the hydrogen bonds are the most important in view of their energy and directionality [7] . Nitrofurantoin (NF) is an antibacterial drug and widely used for the treatment of urinary tract infections [8] , however, a series of potential problems that occur in using such drugs, such as photosensitive [9] , low solubility and low permeability [10] . In recent years, some progresses in improving these insufficient have been made according to crystal engineering through exploring the molecular recognition between NF and co-formers [11] -[13] . Considering the planar structure of NF, the molecules with delocalization П bonds, such as 2,2’-bipyridyl (bipy) and 1,10-phenanthroline (phen), were selected to form cocrystals with NF through molecular recognition. Fortunately, one new polymorphism [14] of NF and two new cocrystals, NF:bipy = 2:1 (2) and NF:phen = 1:1 (3), were obtained. As anticipated, single- crystal X-ray structural analyses of compounds 1-3 revealed the generation of extended supramolecular net- works by means of hydrogen bond interactions, p・・・p interactions and lone pair・・・p interactions.
The Hirshfeld surface [15] [16] provides a remarkable way of exploring intermolecular interactions in molecular crystals using a partitioning of crystal space in a novel visual manner. The surfaces encode information about all intermolecular interactions and offer a facile way of obtaining information on crystal packing. The breakdown of the associated fingerprint plots [17] explores quantitatively the types of intermolecular contacts experienced by molecules and presents this information in a convenient color plot. This plot provides a useful means of revealing significant similarities and differences between related structures by analyzing the packing motifs. The size and shape of the Hirshfeld surface are intimately related to the chemical environment surrounding the molecule, making it ideal for use in comparing different crystal structures incorporating the same molecule. In the context of crystal structure prediction, Hirshfeld surface base tools shows a major advance in analyzing intermolecular interactions [18] [19] and should be considered by the crystal engineers in building molecular architectures. The Hirshfeld surface and associated fingerprint plots of the three compounds have been presented to explore the nature of intermolecular interactions and their relative contributions in building the solid-state architecture. In addition, the thermal stability of the compounds has been investigated associated with the noncovalent interactions observed in the crystal structures.
2. Experimental Section
2.1. Materials and Physical Measurements
Nitrofurantoin, were obtained from Alfa Aesar corporation, ZnCl2, 2,2’-dipyridyl and 1,10-phenanthroline were obtained from Sinopharm Chemical Reagent corporation, all available chemicals were of reagent grade and used without further purification. Elemental analyses were performed on a Thermo 1112 elemental analyzer. Infrared spectra were recorded on a Bruker-Tensor 27 FT-IR spectrometer with the samples prepared as KBr pellets. Thermogrvimetric analyses were performed using a SDT Q600 V8.3 Build101 thermal analyzer at a heating rate of 10˚C/min under an atmosphere of N2.
2.2. Synthesis of Compounds
C8H6N4O5, (1). Nitrofurantoin (β-form, 0.1 mmol, 23.8 mg) and ZnCl2 (0.1 mmol, 13.6 mg) were dissolved in 20 mL MeOH, and stirred about 20 min, followed by slow cooling and evaporation. Yellow needles of 1 were formed after one week (yield 70%). Elemental analysis. Anal. Calcd. (%): C, 40.34; H, 2.54; N, 23.52. Found: C, 40.29; H, 2.55; N, 23.54. IR (KBr, cm−1): 3289 s, 3147 m, 3017 w, 2925 w, 2859 w, 1781 m, 1742 s, 1656 w, 1610 w, 1564 s, 1521 s, 1345 s, 1432 s, 1384.6 m, 1246 s, 1208 m, 1116 s.
C8H6N4O5∙0.5(C10H8N2), (2). Nitrofurantoin (0.1 mmol, 23.8 mg) and 2,2’-dipyridyl (0.1 mmol, 15.6 mg) were dissolved in 15 mL MeCN, and stirred about 15 min, followed by slow cooling and evaporation. Red block-like crystals of 2 were obtained after one day (90%). Elemental analysis. Anal. Calcd. (%): C, 49.37; H, 3.19; N, 22.14. Found: C, 49.40; H, 3.16; N, 22.12. IR (KBr, cm−1): 3162 m, 3101 m, 3019 m, 2923 m, 2855 w, 1790 m, 1741 s, 1656 w, 1615 w,1564 w, 1521 s, 1346 s, 1431 s, 1383 s, 1254 s, 1208 m, 1123 s.
C8H6N4O5∙C12H8N2, (3). Nitrofurantoin (0.1 mmol, 23.8 mg) and 1,10-phenanthroline (0.1 mmol, 18.0 mg) were dissolved in 15 mL MeCN, and stirred about 15 min, followed by slow cooling and evaporation. Olive block-like crystals of 3 were formed after two days (80%). Elemental analysis. Anal. Calcd. (%): C, 57.42; H, 3.37; N, 20.09. Found: C, 57.40; H, 3.39; N, 20.10. IR (KBr, cm−1): 3144 w, 3058 m, 3008 s, 2925 m, 2861 m, 1781 m, 1735 s, 1648 w, 1614 w, 1556 w, 1508 s, 1350 s, 1424 s, 1381 s, 1255 s, 1209 m, 1101 s.
2.3. X-Ray Crystallography
Single crystal X-ray diffraction intensity data were collected using a Bruker SMART APEX CCD diffractometer equipped with graphite monochromated MoKα radiation (l = 0.71073 Å). The data reduction was carried out using the program Bruker SAINT [20] and corrected for absorption using SADABS [21] . The structures were solved by directed methods using SHELXTL [22] and refined by full matrix least-squares on F2 with SHELXL- 97 [23] . Data collection and refinement parameters for compounds 1-3 are summarized in Table 1.
2.4. Hirshfeld Surfaces Analysis
Molecular Hirshfeld surfaces and the associated fingerprint information of 1-3 were calculated using the Crystal Explorer 3.0 software [24] . All bond lengths to hydrogen were automatically modified to typical standard neutron values [25] (C-H = 1.083 Å, N-H = 1.009 Å and O-H = 0.983 Å), while the CIF. files of crystals 1-3 were read into the Crystal Explorer software program for calculations.
3. Results and Discussion
3.1. Crystal Structures Description
Compound 1 crystallizes in monoclinic space group P21/c with β = 109.68˚, which is one new polymorphism of nitrofurantoin [26] [27] . The structural parameters about the polymorphism reported including compound 1 are summarized in Table S1. The selected bond lengths and angles about NF molecule in compounds 1-3 are listed in Table 2, and perspective views of the structures for compounds 1-3 are shown in Figure 1. The bond lengths
![]()
Table 1. Crystallographic data for compounds 1-3.
a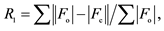 b
b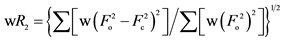 .
.
![]()
Table 2. Selected bond lengths (Å) and angles (˚) of compounds 1-3.
![]()
Figure 1. ORTEP views with atom numbering scheme of compounds 1-3. Thermal ellipsoids are shown at 50% probability level. Symmetry codes for the generated atoms: #A (-x + 1, -y + 1, -z + 1).
and angles of NF molecule in compounds 1-3 are similar, except that the lengths of the C-C bonds in furan ring and the N-O bonds of nitryl are slightly longer in compounds 2 and 3 than those in 1, which might attribute to the weak interactions between NF and bipy (or phen) molecules. As is illustrated in Table S2, the dihedral angle between the furan ring and the hydantoin ring in 3 is 10.98˚, much larger than that in 1 (5.35˚) and 2 (3.26˚), due to the electron repulsion and steric hindrance of phen molecule in 3.
The crystal packing of compound 1 differs from the reported structures of NF [26] [27] , stabilized by combination of N-H・・・O and C-H・・・O hydrogen bonds (Table 3). One imdazole nitrogen atom N1 (x, y, z) acts as donor to O1 atom at (−x + 1, y + 1/2, −z + 1/2), along with C4-H4A・・・O2 (x, y − 1, z), C7-H7A・・・O4 (x, y − 1, z), generating one dimensional parallel ribbon in 1. These hydrogen bonded motif can be described using the graph set notation as  (Figure 2).
(Figure 2).
In 2, one bipy and two NF molecules act as a synthon forming 1D chains with ABB type. One bipy molecule is linked to two NF molecule through H-bond interactions (Table 3), C10-H10A・・・O1 (x, y + 1, z), C13-H13A・・・O2 (−x + 1, −y, −z + 1) and N1-H1A・・・N5 (−x + 1, −y, −z + 1), which give rise to the 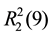 and
and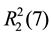 cyclic motifs (Figure 3(a)). Between two NF molecules, there are two H-bond interactions (C7- H7A・・・O5 (−x, −y + 1, −z + 1)), forming a
cyclic motifs (Figure 3(a)). Between two NF molecules, there are two H-bond interactions (C7- H7A・・・O5 (−x, −y + 1, −z + 1)), forming a  motif. The parallel chains stack through p・・・p and nitryl-O・・・p interactions (Table 4 and Table 5) with the average distance of 3.246 Å. The adjecent chains link with the above chain (angle: 28.16˚) through hydrogen bond interactions, C6-H6A・・・O1 (−x + 1, y + 1/2, −z + 1/2) and C11-H11A・・・O2 (x, −y + 1/2, z − 1/2), forming a 3D supramolecular network Figure 3(b) and Figure 3(c).
motif. The parallel chains stack through p・・・p and nitryl-O・・・p interactions (Table 4 and Table 5) with the average distance of 3.246 Å. The adjecent chains link with the above chain (angle: 28.16˚) through hydrogen bond interactions, C6-H6A・・・O1 (−x + 1, y + 1/2, −z + 1/2) and C11-H11A・・・O2 (x, −y + 1/2, z − 1/2), forming a 3D supramolecular network Figure 3(b) and Figure 3(c).
![]()
Table 3. Hydrogen bond parameters (Å, ˚) for compounds 1-3.
![]()
Table 4. Geometrical parameters (Å, ˚) for the π・・・π interactions for compounds 2-3a.
aCg(I) are the centroids of above rings, Ring1 = O3-C5-C6-C7-C8, Ring2 = N5-C9-C10-C11-C12-C13, Ring3 = N5-C18-C17-C16-C15-C19, Ring4 = C12-C13-C14-C15-C19-C20. bCentroid distance between ring I and ring J. cPerpendicular distance of ring centroid I on ring J. dPerpendicular distance of ring centroid J on ring I.
![]()
Table 5. Geometrical parameters (Å, ˚) for the lone pair・・・π interactions for compounds 2-3a.
aCg(I) are the centroids of above rings, Ring2 = N5-C9-C10-C11-C12-C13, Ring4 = C12-C13-C14-C15-C19-C20.
![]()
Figure 2. View of the 1D ribbon-like structure in 1 along the b-axis.
For 3, two NF and two phen molecules link with each other through C-H・・・O and N-H・・・N hydrogen bonds (Table 3), forming an isolated square-planar unit (Figure 4(a)), giving rise to the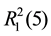 ,
, 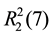 ,
, 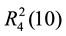 and
and ![]() motifs. The parallel planar units stack through p・・・p and nitryl-O・・・p interactions (Table 4 and Table 5), which are similar to those in compound 2, forming the 1D “square column” motifs (Figure 4(b)).
motifs. The parallel planar units stack through p・・・p and nitryl-O・・・p interactions (Table 4 and Table 5), which are similar to those in compound 2, forming the 1D “square column” motifs (Figure 4(b)).
3.2. Hirshfeld Surfaces Analysis
Herein, the Hirshfeld surfaces of compounds 1-3 are illustrated in Figure 5, showing the surfaces that have been mapped over dnorm and shape-index. The information given in Table 3 is summarized effectively in the Hirshfeld surfaces as the large circular red areas marked with N・・・H and O・・・H, respectively. The other small extents of visible red spots and light-white regions in the dnorm surfaces are indicative of weaker and longer contacts other than hydrogen bonds (Figure 5, dnorm). The hydrogen bonds interactions are also evident in shape-index by a red concave region around the acceptor atom and a complementary blue convex region around the hydrogen bond donor. Apart from hydrogen bonds, the π・・・π interactions stacks for 2-3 are also observed on the same region of the shape index as the pattern of red and blue triangles highlighted by black dashed circles (Figure 5, right).
Meanwhile, the 2D fingerprint plots that decomposed to highlight particular atoms pair close contacts are provided in Figure 6, and the relative contributions of individual intermolecular interactions to the Hirshfeld surfaces area are depicted in Figure 7. In 1, the O・・・H/H・・・O intermolecular interactions appear as a pair of symmetrical large sharp spikes in the fingerprint plots in the region of (1.10 Å, 0.76 Å), which comprise 50.4% of the total Hirshfeld surfaces area. The N・・・H/H・・・N interactions comprise 3.9% of the total Hirshfeld surfaces and represent two small wings with di + de = 3.08 Å, which is longer than the rvdW separation and suggests the absence of any C/N-H・・・N. Furthermore, the H・・・H interactions are displayed in the distribution of scattered points in the fingerprint plots, which spread up to di = de = 1.39 Å and comprise 10.0% of the total Hirshfeld surfaces. Unlike 1, the O・・・H/H・・・O intermolecular interactions are longer in 2 (di + de = 2.27 Å) and 3 (di + de = 2.24 Å), respectively, with the smaller percentage of 45.6% and 36.1% to the total Hirshfeld surfaces, respectively. However, the N・・・H/H・・・N interactions are displayed in two sharp spikes in the fingerprint plots, which spread up to the shorter distance of di + de = 1.98 Å and di + de = 2.16 Å for 2 and 3, respectively, and contribute 8.7% and 13.2% to the total Hirshfeld surfaces, respectively. The H・・・H interactions have more significant contribution to the total Hirshfeld surfaces in 2 (20.2%) and 3 (20.7%) when compared with 1, which spread up to
![]()
Figure 3. (a) View of the hydrogen bond interactions in 2; (b) The 3D grid-like supramolecular structure viewed down the c-axis in 2; (c) A diagrammatic representation of the 3D supramolecular architecture.
![]()
Figure 4. (a) View of the hydrogen bond interactions in 3; (b) A space-filling model of 1D structure in 3.
di = de = 1.08 Å and di = de = 0.9 Å, respectively. The most significant difference amongst 1-3 is the presence of p・・・p/lone pair stacking interactions only in 2 and 3, while introduce the Bipy and Phen molecules. The π・・・π stacking interactions is closer in 3 (di + de = 3.16 Å) than in 2 (di + de = 3.28 Å), and the proportions of the total Hirshfeld surfaces are 4.0% and 2.4%, respectively. The O・・・C/C・・・O contacts associated with lone pair・・・p stacking interactions are closer and maximum in 3 (di + de = 3.08 Å, 7.0%) than that in 2 (di + de = 3.14 Å, 4.7%), respectively.
3.3. Thermal Analysis
The thermal behaviors of compounds 1-3 have been investigated, and the TGA curves are shown in Figure 8. In compound 1, the NF molecule decomposed at 230˚C with the weight loss of 46.2% (calc. 47.4%), which is slightly lower than that of β-NF (240˚C) [28] . For compound 2, the bipy molecule decomposed before the NF molecule at about 154.0˚C with the weight loss of 25.1%, which is in a good agreement with the calculated value of 24.7%. While to compound 3, the NF molecule decomposed before the phen molecule at 175˚C with the weight loss of 26.2% (calc. 27.0%).
4. Conclusion
In summary, the crystal structures, Hirshfeld surfaces and thermal behaviors of the title compounds 1-3 are studied.
![]()
Figure 5. Hirshfeld surfaces mapped with dnorm (front view, left), dnorm (back view, middle) and shape index (right) of NF in compounds 1-3, respectively. The p・・・p stacking interactions are highlighted by black dashed circles.
![]()
Figure 6. Fingerprint plots of compounds 1-3: Full (left) and resolved into N・・・H/H・・・N (middle) and O・・・H/H・・・O (right) contacts, showing the percentages of contacts contributed to the total Hirshfeld surface area of molecules.
![]()
Figure 7. Relative contributions of various intermolecular contacts to the Hirshfeld surfaces area in compounds 1-3.
![]()
Figure 8. TGA curves for 1 (black), 2 (blue), and 3 (violet).
With the construction of hydrogen bond interaction, along with p・・・p/lone pair stacking interactions (for 2 and 3), the supramolecular structures are formed, 1D parallel ribbon structure of 1, 3D grid network of 2, and 1D square column of 3. The Hirshfeld surfaces and 2D fingerprint plots analyses make the understanding of the intermolecular interactions easily. In addition, the thermal stable studies show that the intermolecular interactions play a key role in the thermal properties of the compounds.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (20971101 and 21271142) and the Master Innovation Foundation of Wenzhou University (3160603601011344).
Supplementary Material
Table S1 listing unit cell parameters of nitrofurantoin crystal structures reported in the CSD and in this study. Table S2 listing the dihedral angle between the furan and the hydantoin ring designated by a. Crystallographic data (excluding structure factors) for compounds 1-3 have been deposited with the Cambridge Crystallographic Data Centre (CCDC) as supplementary publication numbers CCDC 1035025, 1035026 and 1035027, respectively. Copies of the data can be obtained on application to CCDC, 12 Union Road, Cambridge CB2 1EZ, UK [Fax: +441223336033; e-mail:deposit@ccdc.cam.ac.uk.
![]()
Table S1. Unit cell parameters of Nitrofurantoin crystal structures reported in the CSD and in this study.
![]()
Table S2. The dihedral angle between the furan and the hydantoin ring designated by a (˚).
NOTES
*Corresponding author.