Adaptability and Stability Analysis of Soybean Genotypes Using Toler and Centroid Methods ()
1. Introduction
The soybean (Glycine max L. Merr.) crop has been grown in a wide range of latitudes in Brazil ranging from 4˚N at Pacaraima―RR to 33˚S at Santa Vitoria do Palmar―RS [1] . Therefore, the main role of soybean breeding programs in Brazil is to develop high-yielding cultivars that are adapted to a variety of growing regions. The understanding of genotype × environment (G × E) interactions will be important for improving the genotypes for higher yields. The presence of G × E interactions in multi-location evaluation trials leads to the production of genotypes that rank among the best in one location although they perform poorly in another. This is the main hampering factor in selecting genotypes with a wide adaptability [2] .
Through a simple joint analysis of variance among G × E trials repeated in two or more environments, the G × E interactions could be detected [3] . Studies of G × E interactions can provide accurate information about each genotype performance and environment variation, if a stability and adaptability analysis may be used [4] .
Among the methodologies for adaptability and stability analysis, those using linear regression have been the most widely used [5] - [7] . However, two characteristics of linear regression methodologies are limiting their use. The first is the difficulty of interpretation when the linearity fails, and the other is that the genotype response pattern is only in one dimension [8] .
In the nonlinear regression method presented by Toler [9] , some improvements were made relating to the linearity response of genotypes and also to the classification of genotypes in distinct groups according to their response patterns. By this method each genotype could show two response patterns. The first is mono-segmented, and it occurs when a single straight line is able to represent the genotype response pattern in favorable and unfavorable environments, whereas the second pattern is bi-segmented and is represented by two straight lines, one demonstrating the genotype response pattern in favorable environment and the other in unfavorable environment.
There is a current trend towards the use of multivariate methods; one of the reasons is the possibility to visualize a genotype performance in a given environment in a dimensional space [8] . Among the multivariate analysis methods, Centroid is based on the dispersion of genotypes and environments in a plane with few axes, and it allows comparisons between genotypes and ideotypes by Cartesian distance. The ideotypes created by this method aim to characterize genotypes with maximum general adaptability and also specific adaptability to favorable and unfavorable environments, as well as identify those with minimum adaptability [10] .
The objective of this study was to assess the adaptability and stability of yield in Brazilian experimental lines of semi-late and late maturing in three locations over two years.
2. Material and Methods
The trials were conducted in 2005/2006 and 2007/2008 growing seasons at 3 locations in Brazil which were at Porangatu―GO, Porto Alegre do Norte―MT and Uberaba―MG. They are part of the Cultivation and Use Value (CUV) network trials of the of the Federal University of Uberlandia breeding program, which aims at improving yield and oil content of soybeans.
Thirteen experimental soybean lines of semi-late/late maturity were evaluated and the commercial cultivar “MSOY-8914” was used as check. The experiments were carried out in randomized complete blocks with 3 replications. The experimental plot consisted of 4 rows of soybean plants of 5.0 meters long, spaced 0.50 meters apart. The two central rows of each plot were harvested, but not the 0.50 meters at each end of the rows. The harvestable area of each plot was 4.0 square meters.
An analysis of variance was carried out for each of the 6 environments (i.e. Porangatu―GO, Porto Alegre do Norte―MT, Uberaba―MG in the 2005/2006 and 2007/2008 growing seasons). Genetic and statistical analysis was performed by the GENES software [11] .
Preceding the joint analyses, variance homogeneity test was performed. This was based on the criterion of the maximum relation between the highest and lowest residual mean squares equaling 7 [4] . Once the residual variances were considered homogeneous, a joint analysis of variance was performed for all environments together. The joint analysis follows the triple design 14 × 3 × 2 (genotypes × locations × years; G × L × Y). The Scott- Knott means grouping test was also performed, with 5% of probability at each location where the genotype effect was observed. The G × L × Y interaction was significant, therefore the analyses of stability and adaptability was performed. First the methodology proposed by Toler [9] was used. This method contained the statistical model to describe the stability and adaptability as follows:
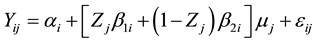 (1)
(1)
where:
Yij: is the average response of genotype i at environment j (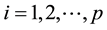 genotypes; and
genotypes; and 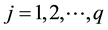 environments);
environments);
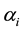 : is the parameter that reflects the value of the response of genotype i in the average productivity environment
: is the parameter that reflects the value of the response of genotype i in the average productivity environment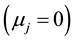 ;
;
 and
and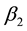 : are parameters that reflect the response sensitivity of genotype i in environments of low
: are parameters that reflect the response sensitivity of genotype i in environments of low 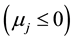 and high
and high 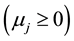 average productivity respectively;
average productivity respectively;
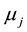 : is the parameter, which reflects the effect of environment j;
: is the parameter, which reflects the effect of environment j;
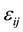 : is the mean experimental error;
: is the mean experimental error;
Zj: is a dummy indicative variable being Zj = 1 when , and Zj = 0 when
, and Zj = 0 when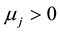 . The estimates
. The estimates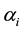 ,
, 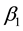 ,
, 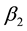 and
and ![]() were calculated by the software Stability [12] . The Toler methodology [9] assumes 5 pattern responses for the genotypes, which are:
were calculated by the software Stability [12] . The Toler methodology [9] assumes 5 pattern responses for the genotypes, which are:
A―convex response and doubly desirable;
B―simple linear response and just desirable at high quality environments;
C―simple linear response does not deviate from average response;
D―simple linear response and just desirable at low quality environments;
E―concave response and doubly undesirable.
These response patterns are assumed according to a hypothesis test based on Student’s t test. First the hypothesis H0: ![]() is tested, which determines whether the model is bi- or mono-segmented. If the result is significant the hypothesis is rejected and Ha:
is tested, which determines whether the model is bi- or mono-segmented. If the result is significant the hypothesis is rejected and Ha: ![]() is accepted; hence it will have two segmented lines. Otherwise H0:
is accepted; hence it will have two segmented lines. Otherwise H0: ![]() is accepted and the model will have only one straight line.
is accepted and the model will have only one straight line.
Therefore, the criteria for classification of genotypes into the 5 groups can be summarized as follows:
![]()
The Centroid method is a multivariate analysis proposed by Rocha et al. [10] , and is based on comparisons of Cartesian distances between genotypes and 4 ideotypes, which are created from experimental data. Later, a modification of this method was proposed by Nascimento et al. [13] ; 3 ideotypes were added in order to improve the sensitivity of methodology in identifying genotypes which have little contribution to the genotypes x environments interaction [14] . Thus, each ideotype and their descriptions according to this method are as follows: Ideotype 1 shows maximum general adaptability; and its values in each environment are represented by the maximum obtained from the group of genotypes studied. Ideotype 2 shows maximum specific adaptability to favorable environments; its values are represented by the maxima in favorable environments and by the minima in unfavorable environments. Ideotype 3 shows maximum specific adaptability to unfavorable environments; its values in favorable environments are represented by the minima and by the maxima in unfavorable environments. Ideotype 4 shows minimum adaptability; its values in each environment are represented by the minima. Ideotype 5 shows mean general adaptability; its values in each environment are represented by the mean obtained in the group of genotypes studied. Ideotype 6 shows mean specific adaptability to favorable environments; its values in favorable environments are represented by the maxima and by the mean in unfavorable environments. The last is Ideotype 7 which is the one of mean specific adaptability to unfavorable environments; its values are represented in favorable environments by the mean and by the maxima in unfavorable environments.
Using the statistical software GENES [11] all analyses of variance as well as adaptability and stability analyses by the Centroid method were carried out. In this method the classification of environments into favorable or unfavorable is made by the environmental index proposed by Finlay and Wilkinson [6] as follows:
![]() (2)
(2)
where:
Yij: mean of genotype i at environment j;
Y: total of all observations;
a: number of environments;
g: number of genotypes.
Thus, the mean Cartesian distance of each genotype to reference ideotypes is calculated by the following formula:
![]() (3)
(3)
where:
Pik: probability that genotype i is similar to ideotype k;
dik: distance from genotype i to ideotype k generated in the Cartesian plane.
3. Results and Discussion
The analysis of variance for each environment was carried out and the results are shown in Table 1. The coefficients of variance (CV) were 14.03% and 21.49% for Porto Alegre do Norte 2005/2006 and Uberaba 2007/2008, respectively. This indicated systematic control of variance sources in the trials, as grain yield is a quantitative character, which was strongly affected by environmental factors.
Analyzing soybean genotypes of semi-early and semi-late maturity Rocha [15] found CVs for grain yield character of 17.95% and 26.04% for the semi-early and semi-late maturity soybeans, respectively. According to Rocha and Vello [16] soybean genotypes of late maturity group are more likely to show higher CV due to their longer exposure to environmental factors.
The relation between the higher and lower residual mean squares of all environments was 2.55. Therefore experimental residual variances were considered homogeneous once the maximum accepted is 7 [4] . Consequently, the joint analysis of variance (Table 2) using the F test could be carried out; notably the interaction effect of G × L × Y was considered significant (P < 0.01). This indicated a differential response of genotypes at different locations [17] . This interaction indicates that the analyzed locations and growing seasons affected the genotypes differently, making it harder to give a general recommendation of cultivars without loss of maximum yield [18] [19] .
The environmental index proposed by Finlay and Wilkinson (1963) was required for both methods used. In the Toler method this index was![]() , and Ij in Centroid. The estimated values are shown in Table 3. The most unfavorable environment was Porangatu 2005/2006 followed by Uberaba 2005/2006 and Porto Alegre 2007/2008.
, and Ij in Centroid. The estimated values are shown in Table 3. The most unfavorable environment was Porangatu 2005/2006 followed by Uberaba 2005/2006 and Porto Alegre 2007/2008.
![]()
Table 1. Average grain yield (kg∙ha−1), mean square error (MSE) and coefficient of variance (CV) of soybean lineage trials, of semi-late/late maturity at six environments (2 growing seasons and 3 locations).
![]()
Table 2. Joint analysis of variance of grain yield (kg∙ha−1), of 14 soybean genotypes of semi-late/late maturity at three locations across two growing seasons.
*, **: significant at 5% and 1% levels of probability, respectively.
![]()
Table 3. Location classification using an enviromental índex of soybean genotypes of semi-late/late maturity in three loca- tions across two growing seasons.
Therefore, all of them presented negative environmental indices. On the other hand the most favorable environment was Porto Alegre do Norte 2005/2006 followed by Porangatu 2007/2008 and Uberaba 2007/2008. Consequently, all locations showed positive environmental indices.
The adaptability and stability analysis according to the Toler method assumed 5 possible groups for each ge-
notype. The estimated parameters![]() ,
, ![]() ,
, ![]() ,
, ![]() and
and ![]() which were necessary for this classification are presented below (Table 4). By analyzing results for
which were necessary for this classification are presented below (Table 4). By analyzing results for ![]() parameter, we noted that the majority of genotypes are not significant (H0:
parameter, we noted that the majority of genotypes are not significant (H0:![]() ). Therefore, they had linear response patterns, which means that
). Therefore, they had linear response patterns, which means that
their behavior in favorable and unfavorable environments was similar, hence they classified in Groups B, C or D.
However, 6 genotypes presented significant results for ![]() parameter (Ha:
parameter (Ha:![]() ), thus they showed
), thus they showed
nonlinear pattern responses which resulted in a bi-segmented model. Notably, the genotypes UFU-001, UFU-003, UFU-0010, and UFU-0011 were classified in Group A which means that they were well adapted to high quality environments (e.g. high soil fertility, controlled biotic and abiotic stresses), and they needed these types of environment to express their maximum yield potential [20] .
Also in the bi-segmented model were the UFU-008 and UFU-003 genotypes, however they were also classified in Group E, for good performance in the most unfavorable environments, but a poor in the most favorable. A single straight line was able to explain the behavior of Groups B, C and D of genotypes. In order to have the
![]()
Table 4. Mean yield (Ῡ.i.) (kg∙ha−1), Toler’s estimated parameters![]() , and response pattern (group) for each geno- type according to this method (2 growing seasons and 3 locations).
, and response pattern (group) for each geno- type according to this method (2 growing seasons and 3 locations).
*, **: significant at 5% and 1% levels of probability, respectively, through student’s t test.
genotypes classified in these groups the estimates for ![]() parameter were used. If this is equal to one, as
parameter were used. If this is equal to one, as
we had for UFU-002, UFU-004, UFU-005, UFU-007, UFU-009 and MSOY-8914 genotypes, they were classified in Group C of general adaptability. This group is characterized by good plasticity and capability to adjust to environment variations [20] [21] .
For those genotypes whose estimates for the ![]() parameter were significantly higher than one (e.g. UFU-006), classification was in Group B of specific adaptability to high quality environments. Finally, if estimates for the
parameter were significantly higher than one (e.g. UFU-006), classification was in Group B of specific adaptability to high quality environments. Finally, if estimates for the ![]() parameter were significantly lower than one, the genotypes were classified in Group D
parameter were significantly lower than one, the genotypes were classified in Group D
of specific adaptability to unfavorable environments.
These response patterns can be better visualized in Figure 1 and Figure 2 which allow comparisons between the behaviors of genotypes UFU-0010 and UFU-001 classified in Group A, and genotypes UFU-0013 and UFU-008 classified in Group E. Analyzing the Group A genotypes in Figure 1 and Figure 2; the convex pattern is clear as both UFU-0010 and UFU-001 genotypes showed good tolerance in environments with performance below the average, but efficiently responded to improving environmental quality. In fact, for these genotypes we found seed yield increases of about 1800 kg∙ha−1 when comparing their yields in the average environment and in the most favorable environments, respectively. A distinct concave pattern response is shown by UFU-0013 and UFU-008 in Figure 1 and Figure 2. They showed satisfactory yields in the most unfavorable environments. However, these genotypes showed yield losses in favorable environments. A possible reason for that is they were unable to respond advantageously to the improvement of environmental resources as did the genotypes classified in group A. The genotypes, which were classified in Group E, could meet low-technology farming requirements. However, those of Group A are more recommended as they did not seem to have a stabilization of yield increase while improving environmental quality [20] .
In Figure 3 the response pattern of Groups B, C and D is illustrated. The genotype UFU-006 was characterized by its high response in favorable environments. It had a yield increase of nearly 1250 kg∙ha−1 when comparing its yield in mean and in the most favorable environments. By analyzing UFU-0012 genotype performance (Figure 3), it was possible to infer superior yields in unfavorable environments compared to other genotypes. However, its yield response in favorable environments was close to zero.
![]()
Figure 1. Soybean genotype response of semi-late/late maturity classified in bi-segmented groups, pattern A (UFU-0010) and pattern E (UFU-0013), evaluated in 2005/2006 and 2007/2008 growing seasons in three locations.
![]()
Figure 2. Soybean genotype response of semi-late/late maturity classified in bi-segmented groups, pattern A (UFU-001) and pattern E (UFU-008), evaluated in 2005/2006 and 2007/2008 growing seasons in three locations.
![]()
Figure 3. Soybean genotype response of semi-late/late maturity classified in mono-segmented groups (B, C and D pattern), evaluated in 2005/2006 and 2007/2008 growing seasons in three locations.
Centroid adaptability and stability analysis differ from other methods by considering genotypes with maximum specific adaptation, those with maximum yields in a certain group of environments (favorable or unfavorable) and minimum yields in another group [10] .
Table 5 shows the genotype classification according to Centroid method. It was found that 78.5% of genotypes were classified in Ideotype V, which means genotypes with mean general adaptability. Genotype MSOY- 8914, which was used as control, stands out as well as UFU-007 that obtained the highest probability (0.3369) of belonging to Ideotype V. This group of genotypes may have the lowest contribution to the G × E interaction; therefore they are considered the most stable [14] [22] .
Notably UFU-002 genotype was the only one classified in Ideotype VI. The response pattern in this group considered maximum yields in favorable environments and the mean yield in unfavorable ones. This Ideotype was a modification of the original method proposed by Rocha et al. (2005) and it allowed a more biological sense to the analysis as the addition of intermediate ideotypes avoids comparisons with extreme genotypes [13] . Preceding the graphical analysis of genotypes dispersion, a principal component analysis as shown in Table 6 was carried out. Based on values from Table 6, the first two principal components could explain 62.82% of the total variation. Similar values were obtained by Barros et al. [23] , when evaluating the adaptability and stability of 29 soybean genotypes in 6 environments. They reached 67% of total variation explained by the first two components.
Analyzing Figure 4, it is possible to quickly relate the genotypes to each ideotype. The arrow shape created by the connection between points, which was representing ideotypes, allows a quantitative interpretation of genotypic adaptability. As genotypes move away from the tail to the arrow tip it is assumed that productivity increases gradually. Another similar interpretation can be made relating to the arrow’s central axis; those located above are more adapted to favorable environments, and those below more adapted to unfavorable environments. Also, Figure 4 shows a visibly greater concentration of points near Ideotype V, meaning that the majority of genotypes were of mean general adaptability.
Using Centroid method with 4 ideotypes Oliveira et al. [24] concluded that 44% of the 16 genotypes were classified in Ideotype I of maximum general adaptability. However, Marques et al. (2011) [25] using Centroid method proposed by Nascimento et al. (2009) found that 43% of the 7 evaluated genotypes were classified in Ideotype V. By this method, the genotype UFU-002 stood out as being nearest to the arrow tip. Furthermore it is located above the arrow central axis, therefore it can be inferred that this was a genotype with good adaptability, mainly to favorable environments.
![]()
Figure 4. Dispersion of the 14 soybean genotypes into the two first principal components coordinate system considering yield (kg∙ha−1) across two growing seasons and at three locations. The seven points which are numbered with Roman numerals represent the ideotypes where; I, maximum general adaptability; II, maximum specific adaptability to favorable environments; III, maximum specific adaptability to unfavorable environments; IV, minimum adaptability; V, mean general adaptability; VI, mean specific adaptability to favorable environments; VII, mean specific adaptability to unfavorable environments.
![]()
Table 5. Establishment of ideotypes for soybean genotypes of semi-late/late maturity evaluated in two growing seasons in three locations through Centroid’s method.
IV: minimum adaptability; V: mean general adaptability; VI: mean specific adaptability to favorable environments; VII: mean specific adaptability to unfavorable environments.
![]()
Table 6. Cumulative and proportion explained by each principal component.
4. Conclusion
The genotype UFU-008 was classified by the Toler and Centroid methods as being of low adaptability capacity. An agreement was also reached by both methods relating to the genotypes UFU-005, UFU-007, UFU-009 and MSOY-8914. They were classified as being of general adaptability and unaffected by G × E interactions. The mean grain yield of the genotypes UFU-001, UFU-002, UFU-006, UFU-0010, and UFU-0011 was above Brazil’s soybean mean yield in the 2012/2013 growing season (2939 kg∙ha−1). As such, these genotypes may be adapted to high quality environments where they are likely to express their maximum yield potential.
Acknowledgements
This study was partially funded by a scholarship from the Brazilian Coordination for the Improvement of Higher Education Personnel (CAPES―Process BEX 11900/13-0) and supported by the Brazilian National Council for Scientific and Technological Development (CNPq) and by the Research Support Foundation of Minas Gerais State-Brazil (FAPEMIG).