High-throughput simple sequence repeat (SSR) markers development for the kelp grouper (Epinephelus bruneus) and cross-species amplifications for Epinephelinae species ()
1. INTRODUCTION
Groupers (family Serranidae, subfamily Epinephelinae) comprising about 159 marine fish species in fifteen genera are the most intensively exploited group in marine fishing [1,2]. Groupers are also in demand as new aquaculture species in East and South-East Asia because of their high market price, rapid growth and adaptability to high density, rearing conditions [3,4]. The high market price has led to overexploitation of these fish and increased aquaculture. Among the subfamily Epinephelinae, the genus Epinephelus is one of the largest genera among bony fish with 98 species including many economically important species for both capture fisheries and aquaculture [1,5]. The kelp grouper (Epinephelus bruneus) is one of the most economically important species in Japan. However, it has been listed as Vulnerable in the IUCN Red List Category & Criteria (www. iucnredlist.org) due to the sharp decline in catch. Although aquacultural production of kelp grouper has been attempted in response to the decreases in natural resources and increases in price, analysis of the genomics of this species has rarely been explored.
Simple sequence repeat (SSR) has been particularly valuable in parentage analyses, population genetics, conservation or management of biological resources and gene mapping because of their ease of use, co-dominance and high levels of polymorphism [6,7]. Relatively large numbers of SSR markers have been recently developed and characterized for grouper species e.g., Plectropomus maculatus [8], E. lanceolatus [9,10], E. fuscoguttatus [11], Mycteroperca tigris [12], Cromileptes altivelis [13]. However, there has been only one published research study on the SSR primers for E. bruneus in which dinucleotide markers are developed [14]. Although dinucleotide SSR markers are powerful tools for parentage analyses and gene mapping, analyses are frequently complicated by the presence of stutter bands caused by polymerase slippage during PCR amplification. This results in secondary products containing one or more repeat units less than the primary allelic band. The stutter bands are sometimes equivalent to the intensity of the primary band which decreases the accuracy of the genotypic characterization, particularly in population genetics studies [6]. To overcome this problem, tri-, tetraor more nucleotide SSR which possess fewer problems with stutter bands should be used, however, these SSR markers are much less abundant in the genome compared to dinucleotide SSR markers. Recently developed next-generation sequencing (NGS) technologies have allowed reassessment of the technical aspects of SSR search techniques [15]. A notable merit of NGS-based SSR search technique is that various repetitive elements can be explored in shotgun libraries, which enables simultaneous search for nondi-nucleotide SSR markers without implementing special procedures for SSR-enrichment [16]. In human forensic DNA, a consensus was reached that tetra-nucleotide repeat markers should be used as the gold standard for individual identification [17]. Currently, only tetraand penta-nucleotide SSR markers are acceptable for routine human forensic casework [18].
SSR markers can be transferable from different genotypes within or between species or between genera [19]. Such interspecific or intergeneric transferability makes SSR markers a useful tool for genetic studies, such as fingerprinting, genetic mapping, and molecular marker identification. In the present study, we report the development of 1290 SSR markers from whole genome sequences of E. bruneus using 454 pyrosequencing to obtain SSR markers for addressing both population genetics studies and gene mapping. In addition, the developed SSR markers were used to conduct amplification tests with five grouper species i.e., E. bruneus, Hyporthodus septemfasciatus, P. leopardus, E. lanceolatus and E. coioides in order to characterize cross-species polymorphism as well as find SSR motifs that have potential as a highly versatile SSR marker in groupers (Epinephelinae).
2. MATERIALS AND METHODS
2.1. Whole Genome Shotgun Sequences Assembly
Large whole genome shotgun (WGS) sequences were generated from wild kelp grouper. A quarter plate of DNA sequencing was performed with 454 pyrosequencing on a Genome Sequencer FLX-454 System (GS FLX sequencer). Sample preparation and DNA sequencing was performed according to the manufacturer’s instructions (Roche Diagnostics, Mannheim, Germany). The raw reads from the GS FLX454 were assembled using Newbler software version 2.3 (Roche Diagnostics) and the WGS contigs and singletons were generated. Briefly, the adapters and poor sequence data were removed by using the built-in adapter removal tools in Newbler. After sequence trimming and clean-up, de novo assembly was performed using the default parameters. The sequences of contigs and singletons were used for SSR identification.
2.2. SSR Identification
A pipeline program, Auto-primer [20], which automatically runs two software programs i.e., Tandem Repeats Finder ver. 4.0.4 [21] and Primer3 ver. 2.2.2 beta [22], was used to identify sequences containing repeat motifs (di-, tri-, tetraand penta-nucleotide) and to predict their appropriate PCR primers from WGS contigs and singletons. In Tandem Repeats Finder, alignment weights for match, mismatch and indels were set as two, seven and seven, respectively. Matching and indel probabilities were set as 0.8 and 0.1, respectively. Since nonunique repeat motifs such as reverse-complement repeat motifs (e.g., AC and GT) and translated or shifted motifs (e.g., AAT, ATA, ATT, TAA, TAT and TTA) were grouped together [23], a total of three unique di-nucleotide repeats, ten unique tri-nucleotide repeats, 25 unique tetra-nucleotide repeats and 47 unique penta-nucleotide repeats were obtained from the kelp grouper genome. SSR markers containing genomic DNA sequences with flanking sequences greater than 30 bp on either side of SSR markers were collected for primer design.
2.3. SSR Marker Development
In order to adjust the PCR product size, primers were redesigned using web software WebSat, which is accessible through the Internet [24]. The thresholds for di-, tri-, tetra-, penta-nucleotide repeat minimum were set as six in all motif length. The parameters were employed to design primers except for the product size was set as 100 - 250 bp. Each forward primer was 5’ labeled with fluorescent dye: Tetrachloro-fluorescein (TET).
2.4. Sample Collection and Polymorphism Test
To examine SSR markers, newly-designed PCR primer pairs were tested for amplification using the samples from eleven kelp groupers. Those fish are already known the relationship between the parental fish and progenies. Three individuals each of H. septemfasciatus, P. leopardus, E. lanceolatus and E. coioides were used to characterize cross-species amplification from wild resources. These species were chosen as they are of economic and current research importance.
2.5. SSR Marker Validation
PCR amplification of the SSR markers was performed on MJ PTC-100 thermal cycler (Bio-Rad, USA) in 11 μl reaction volume containing 0.5 pmol/μl of unlabeled primer, 0.05 pmol/μl of fluorescence-end-labeled (TET) primer, 1× Ex Taq buffer, 2.0 mM MgCl2, 0.2 mM dNTP, 1% BSA, 0.25 U of Taq DNA polymerase (TaKaRa: Ex-Taq) and 50 ng template DNA. Suitable annealing temperatures for each SSR marker were used. PCR conditions were 95˚C for 5 min for initial denaturation, followed by 36 cycles at 95˚C for 30 s, 56˚C for 1 min, 72˚C for 1 min, with a final extension at 72˚C for 10 min. Amplification products were mixed with an equal volume of loading buffer (98% formamide, 10 mM EDTA (pH 8.0), 0.05% bromophenol blue), heated for 10 min at 95˚C and then immediately cooled on ice. The mixture was loaded onto 6% PAGE-PLUS gel (Amresco, OH, USA) containing 7 M urea and 0.5× TBE buffer. The PCR products were visualized using FMBIO III MultiView fluorescence image analyzer (Hitachi-soft, Tokyo, Japan). SSR markers that showed more than one alleles per locus were recognized as polymorphic markers.
3. RESULTS AND DISCUSSION
3.1. 454 Sequencing Result
The raw sequence data from a quarter-plate run of the 454 sequencing yielded 84.1 Mbp containing 213,073 reads or sequences with an average length of 395 bp (maximum: 677 bp, minimum: 40 bp) (Table 1). A total of 10,766 reads (approximately 5.1%) were assembled into 4551 contigs with an average length of 385 bp (maximum: 6510 bp, minimum: 100 bp), leaving 166,867 singletons. The mean length of these 171,418 sequences (4551 contigs plus 166,867 singletons) was 392 bp, which was similar to that of the raw sequences. This number also agreed with the results from previous studies on 454 sequencing run in nonmodel species including the bream, Megalobrama pellegrini, with average read length of 404 bp [25] and the abalone, Haliotis diversicolor supertexta, with average length of 385 bp [26]. To our knowledge, this is the first large scale study

Table 1. Sequencing statistics using 454 sequencing platforms.
of genomic data from E. bruneus.
3.2. SSR Loci Isolation
Of the 171,418 unique sequences, 2348 (1.37%) sequences comprising 1118 (47.6%), 488 (20.8%), 473 (20.1%) and 269 (11.5%) of di-, tri-, tetraand pentanucleotide repeat motifs, respectively, were suitable for primer design (Table 2). In general, the number of repeats decreased with motif length. Of 2348 sequences, the average number of repeats with di-, tri-, tetraand penta-nucleotide repeat motifs were eighteen, twelve, ten and eight, respectively. Currently, CAG and AGAT repeat types predominate in vertebrate SSR markers while di-nucleotide (CA) repeats are the common SSR markers developed for genetic studies in fish [27, 28]. The most common repeat motifs of di-, tri-, tetraand pentanucleotide in E. bruneus included AC/GT (93.3%), AAT/ ATT (37.5%), ACAG/CTGT (13.7%) and AAAAT (12.3%), respectively. Although the relative frequency of SSR motif types was different among the species, AC/GT, AAT/ATT and AGAT/ATCT were found as the common repeat motifs with a high percentage of loci suitable for primer design in the North American fish (Etheostoma okaloosae) [29]. These SSR motifs are also abundant in the E. bruneus genome.
3.3. SSR Marker Development and Validation
A total of 1466 primer sets were redesigned from 2348 sequences using the web software WebSat (Table S1) of which four types of SSR markers i.e., di-, tri-, tetraand penta-nucleotide repeat motifs were included with the primer numbers of 826 (56.3%), 317 (21.6%), 254 (17.3%) and 69 (4.7%), respectively. Since the threshold for all repeat minima was set as six, 71 penta-nucleotide repeat motifs (26.4% of identified penta-nucleotide repeat motifs from the kelp grouper genome) were ignored as SSR marker in this study. Consequently, only 25.7%

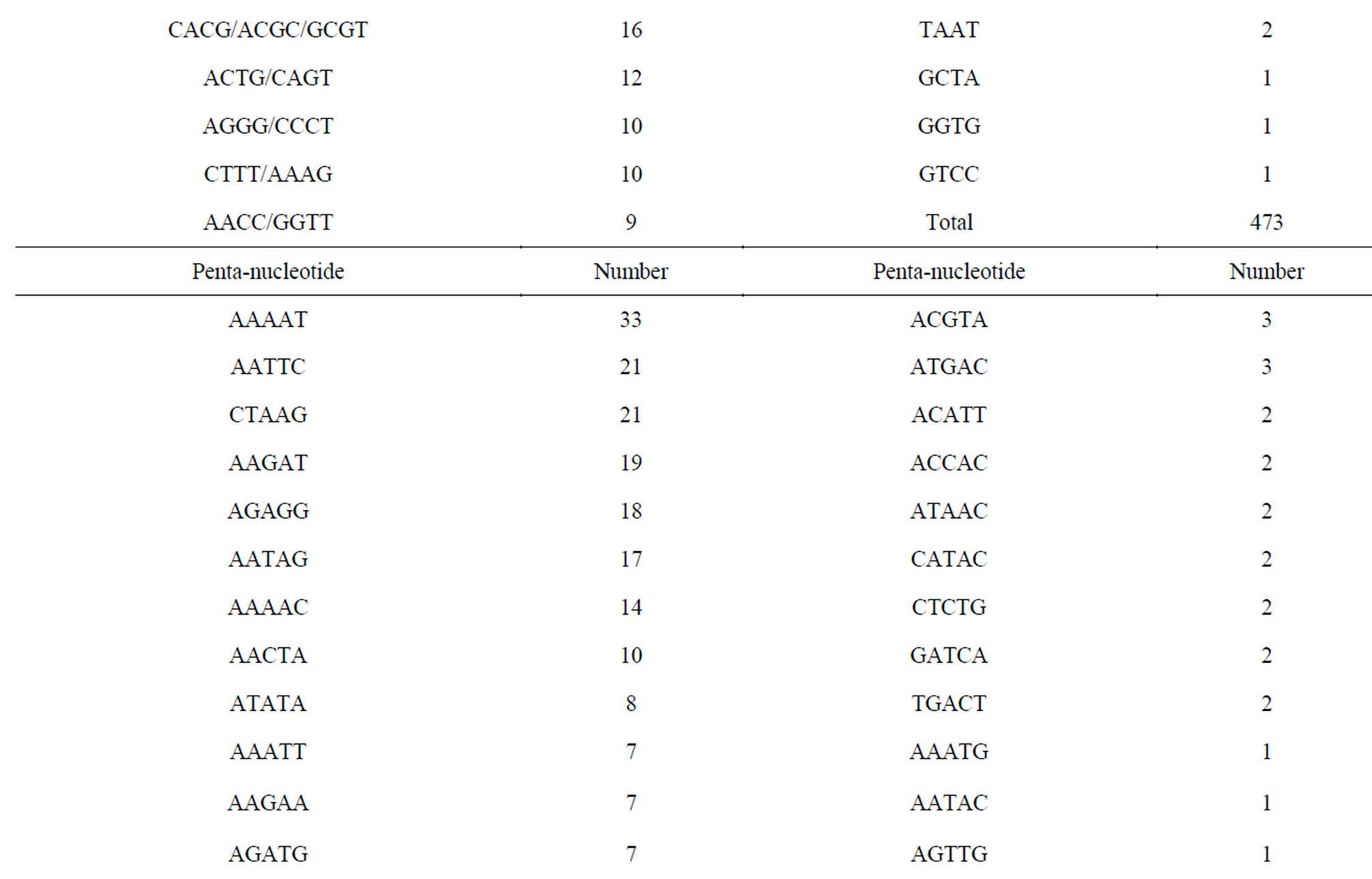

Table 2. Summary of SSR motifs with suitable for primer design of kelp grouper.
of identified penta-nucleotide repeat motifs from kelp grouper genome could be used for redesigning the primer. Newly-designed primers were tested in the eleven kelp groupers. Among the primer sets tested, 1244 primer sets produced strongly amplified expected fragment size, of which 905 (72.7%) primer sets showed clear amplification with polymorphic patterns (Table 3).
The amplified and polymorphic marker results for each nucleotide repeat motif are summarized in Tables 4(a) and (b). In each of diand tri-nucleotide repeat motifs, there are only slight differences between proportion of amplification and polymorphism among species. It seems that each diand tri-nucleotide repeat motif has a similar potential for amplification to polymorphic marker conversion ratio. ACAG, AGAT, ATCC, AATC and AAAC repeat markers accounted for 61.5% among the successfully amplified 20 tetra-nucleotide SSR motif markers, of which ACAG, AGAT and ATCC repeat markers totally accounted for more than 50% of the polymorphism in E. bruneus. In addition, AGAT and ATCC repeat markers exhibit high amplification to polymorphic marker conversion ratio (more than 90%). A/T-rich motif, containing A or T nucleotides > 50%, accounted for 69.6% of polymorphic tri-nucleotide SSR markers, while G/C-rich motifs, containing G or C nucleotides > 50%, were 30.4% of polymorphic tri-nucleotide SSR markers. Similarly, in polymorphic tetraand penta-nucleotide SSR markers, the A/T-rich SSR markers were the vast majority (more than 82%) compared with G/C-rich SSR markers in E. bruneus.
Prior to our study, twelve SSR markers had been developed in E. bruneus using the traditional method [14], and they reported on development of dinucleotide markers. Di-nucleotide SSR markers, while providing powerful discrimination, do not provide high-precision
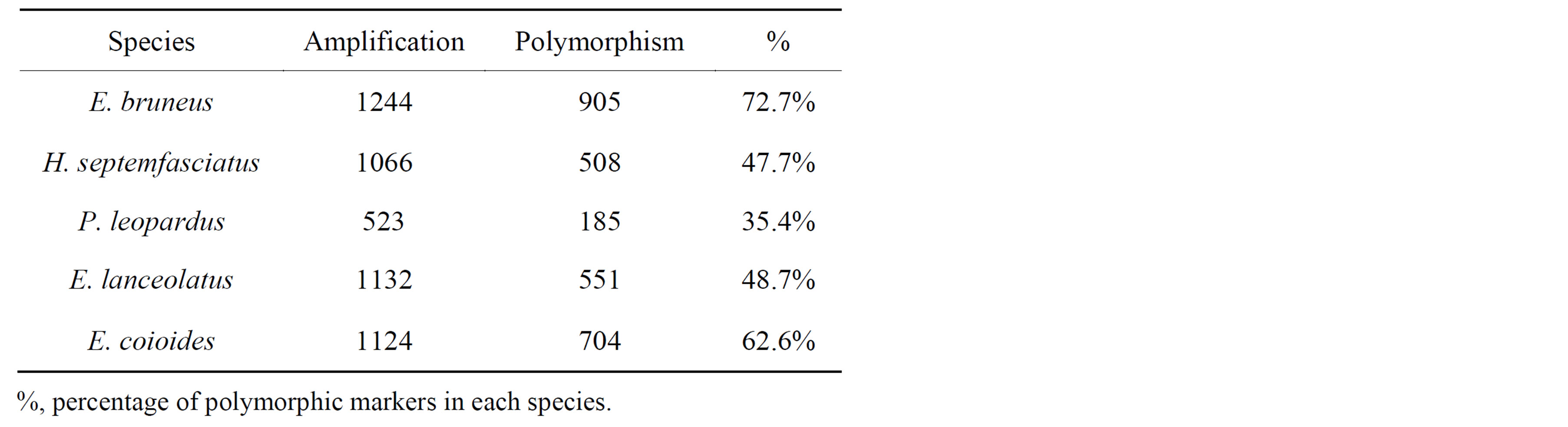
Table 3. Statistics of SSR markers developed in E. bruneus.
genotyping needed for comparative multi-locus profiling. In this study, 905 polymorphic SSR markers were developed using 454 pyrosequencing of which nondinucleotide repeat motifs including 184 tri-, 176 tetraand 52 penta-nucleotide markers were successfully developed. Although unclear amplification or monomorphic pattern was obtained for 561 loci, a comparably high primer to polymorphic marker conversion ratio (62%) was achieved which was similar to that observed in the mottled skate (Raja pulchra) [30]. Of the 1244 primer sets, a total of 742 (59.6%) were successfully validated as polymorphic markers in the future mapping family (Table S2). The polymorphic SSR markers were genotyped in the mapping panel for a future linkage map project.
3.4. Characterization of Cross-Species Amplification
The cross-species amplification of the 1466 SSR loci were conducted in an additional four species, H. septemfasciatus, P. leopardus, E. lanceolatus and E. coioides using three individuals for each species. These grouper species are economically important for both capture fisheries and aquaculture. The number of SSR loci which
were successfully amplified from H. septemfasciatus, P. leopardus, E. lanceolatus and E. coioides were 1066, 523, 1132 and 1124 SSR markers, respectively. Of these, 508 (47.7%), 185 (35.4%), 551 (48.7%) and 704 (62.6%) SSR markers, respectively, showed specific polymorphic products (Table 3).
There are some of the loci possessing unclear amplification or monomorphic pattern in E. bruneus. Although unclear amplification is lacking available loci for the species tested, monomorphism markers are likely to be due to the critically small sample size. In addition, it has been reported that sequences within protein-coding regions generally show lower levels of polymorphism due to functional selection pressure [7]. Hence, a total of 1290 SSR loci (671 di-, 307 tri-, 245 tetraand 67 pentanucleotide) that produced amplified products in more than one species were lodged with the DDBJ database (Accession numbers: AB755818 - AB757107). The SSR locus ID, sequence, sequence length, repeat type, number of repeats, repeat position, primer sequences, primer position and expected PCR fragment size are summarized in the supplement file (Table S1). BLAST search was conducted to compare each of the 1290 SSR loci against the database for ‘nucleotide collection (nr/nt)’ optimized for highly similar sequences. As the result of the BLAST searches (E value < e−48), although fourteen SSR sequences identified from other grouper species including E. akaara (one SSR locus), E. corallicola (one SSR locus), E. fuscoguttatus (six SSR loci), E. lanceolatus (two SSR loci) and the hybrid between E. coioides and E. lanceolatus (four SSR loci) were not novel loci, the sequence composing SSR markers identified in our study did not match with the published sequences for E. bruneus in the database. In contrast, the sequence homologies are indicative of close evolutionary relationships between E. bruneus and other species of the genus Epinephelus. Results indicate that we successfully developed new SSR markers for E. bruneus. However, it should be noted that the number of E. bruneus sequences currently available in the NCBI database is still limited (twelve SSR loci are available) [14].
As shown in Tables 4(a) and (b), the frequencies of amplification or polymorphism among the additional four species are similar to E. bruneus except for that of P. leopardus. This result is probably a reflection of the different genera between Epinephelus and Plectropomus [31]. ACAG, AGAT and ATCC repeats accounted for more than 40% of the successfully amplified tetra-nucleotide SSR motifs among the four species tested, which is not markedly different from that of E. bruneus (44.4%). These three motifs totally have more than 50% of the polymorphism. AGAT repeats exhibit relatively high amplification to polymorphic marker conversion ratio expect for P. leopardus. Also, SSR polymorphism is based on size differences due to varying number of repeat units contained by alleles at given locus [6]. These results suggest that sequence composed AGAT repeat motif is conserved, high SSR mutation rate and could be used as a target for SSR marker development for the genus Epinephelus. Although we could not find a unique motif in the genus Plectropomus, there are different A/Trich and G/C-rich frequencies between Epinephelus and Plectropomus. G/C-rich motifs accounted for 51.5% of the successfully amplified tri-nucleotide repeat motifs in P. leopardus. This is relatively high percentage compared with E. bruneus (32.0%), H. septemfasciatus (34.5%), E. lanceolatus (34.8%) and E. coioides (35.1%). Similarly, in successfully amplified tetraand pentanucleotide SSR markers, the G/C-rich SSR markers were the higher percentage in comparison to A/T-rich SSR markers (about 1.43 - 2.18 times).
Although the cross-species transferability of SSR markers is unevenly distributed among taxa, over 40% of polymorphic marker transfers have been observed in different genera of fish within the same family, and 25% of families within the same order [19]. In our case, among the 905 SSR markers polymorphic in E. bruneus, only 15.0% were polymorphic in P. leopardus even though this species also belongs to the same family (Table 5). The sample size for recognition of marker polymorphism was limited, which probably reflects the rela-
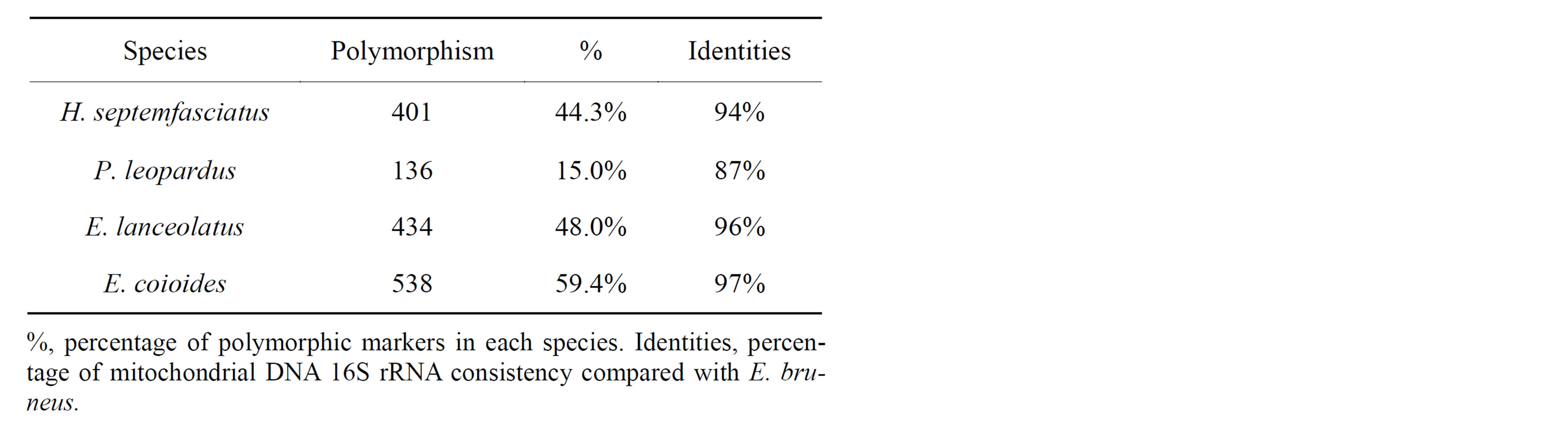
Table 5. Statistics of polymorphic SSR markers in E. bruneus in related-species.
tively low transferability of polymorphic SSR markers. Cross-species amplification success is only achieved when the primer sequences are conserved between species and there is a decrease in the number of amplified loci with the increase in divergence between species [32]. It has been reported that a significant negative correlation was found between genetic divergence (based on mitochondrial DNA 16S rRNA) and SSR transferability from multiple cross-species amplification studies in Sparidae [33]. In order to compare the sequences with differences of mitochondrial DNA, the alignment of 16S rRNA gene sequences between E. bruneus (AY947562) and H. septemfasciatus (AY947559), P. leopardus (AF297298), E. lanceolatus (AY947588) or E. coioides (AY947608) were conducted using BLASTN. Results indicated that comparably high transferability is observed when the genetic divergence of 16S between E. bruneus and each tested species is less than 5% (Table 5) which is in accordance with the results of Carreras-Carbonell et al. [34]. These results support the phylogenetic tree of groupers based on DNA sequence data from two mitochondrial and two nuclear genes [31]. Considering this phylogenetic tree of the groupers, the genera Epinephelus, Mycteroperca, Hyporthodus may have a sequence composing AGAT motif repeat as a conserved region and polymorphism and the sequence comprise G/C rich motif might be an important parameter regarding grouper evolution.
Additionally, 40 SSR markers showed polymorphism in all the other species we tested (Table S3). The results suggested that these markers have a possibility to be used as universal markers for groupers. Interestingly, there are three SSR markers composing the AGAT SSR motif which accounts for 42.9% of the nondi-nucleotide markers among the 40 SSR markers (33 di-, 1 triand 6 tetra-nucleotides). Although we could not find a unique motif in the genus Plectropomus, the AGAT SSR motif has a high potential as a highly versatile SSR marker in grouper Epinephelinae. These results suggest that the AGAT SSR motif might be a target for further screening of larger sets of SSR markers. However the number of genera and number of samples for each genus are limited, the 1290 SSR markers (671 di-, 307 tri-, 245 tetraand 67 penta-nucleotides) developed in this study will be useful for future genetic studies such as parentage analyses, population genetics, conservation or management of biological resources and gene mapping in groupers (Epinephelinae).
4. CONCLUSIONS
The pyrosequencing method was applied to develop dito penta-nucleotide markers for addressing population genetics studies and gene mapping for kelp grouper (E. bruneus). A total of 213,073 raw reads were obtained and 171,418 unique sequences were generated with an average length of 392bp, of which 2348 (1.37%) sequences contained SSR motifs which were suitable for primer design. AC, AAT, ACAG, AGAT and ATCC were found as the common repeat motifs in kelp grouper. A total of 1466 primer sets were designed from 2348 sequences. Among 1466 SSR markers, 1244 primer sets produced strong PCR products, of which 905 (72.7%) were polymorphic in kelp grouper. A relatively high ratio of primer to polymorphic marker conversion (62%) was achieved by this method. In the cross-species amplification, over 40% of the markers amplified specific polymorphic products in the fish belonging to the same subfamily including H. septemfasciatus, E. lanceolatus and E. coioides; however, only 15% were polymorphic in P. leopardus. G/C-rich motifs accounted for 51.5% of successfully amplified tri-nucleotide repeat motifs in P. leopardus, which was relatively high percentage compared with E. bruneus (32.0%), H. septemfasciatus (34.5%), E. lanceolatus (34.8%) and E. coioides (35.1%). ACAG, AGAT and ATCC repeats accounted for more than 40% of the successfully amplified tetra-nucleotide SSR motif, and these nucleotide repeat motifs exhibited more than 50% of polymorphism in all species tested. In addition, there are three SSR markers containing the AGAT SSR motif (42.9% of nondi-nucleotide markers) among the 40 SSR markers (33 di-, 1 triand 6 tetra-nucleotide) that showed polymorphism in all the species we tested. Results indicate that the AGAT SSR motif has a high potential as a highly versatile SSR markers in groupers since the genus Epinephelus is the largest group among grouper Epinephelinae. The SSR markers developed in this study can be employed to obtain reliable genetic variability estimates for groupers (Epinephelinae).
ACKNOWLEDGEMENTS
This research was supported by JST/JICA, SATREPS (Science and Technology Research Partnership for Sustainable Development). We would like to thank Kishiko Kubo, Dr. Kanako Fuji and Dr. Eriko Koshimizu for their significant support and contributions during the whole experiment.
NOTES
#Corresponding author.