1. Introduction
Virus-Host interaction has been proven to be important in the last decade. Indeed, the cells rely on their restriction factors to protect them from viral infections, while viruses like HIV count on the cells’ proteins and cellular pathways to replicate [1] . Host factors such as the C-C chemokine receptor type 5 (CCR5) have led to discovering the Maraviroc used as an antiretroviral medication in the United States [2] . In contrast, other factors like C-X-C Motif Chemokine Receptor 4 (CXCR4) and the Apolipoprotein B mRNA editing enzyme catalytic polypeptide-like 3G (APOBEC3G or A3G) are potent host defense factors that are still under investigation [3] [4] . APOBEC3G (Apolipoprotein B mRNA editing enzyme catalytic polypeptide-like 3G) acts as a host restriction factor by deaminating cytidine (C) to uridine (U) in the negative strand of the HIV sequence and consequently changing guanine (G) into adenine (A) on the positive strand, causing the G to A hypermutation in the absence of the viral infectivity factor (Vif) [5] . The host factor comprises eight (8) exons [6] . APOBEC3G can also act by blocking steps in reverse transcription and viral integration into the host genome [7] . HIV responds to the antiviral activity of APOBEC3G by targeting it to degradation using Vif. Nevertheless, APOBEC3G antiviral activity remains a possible therapeutic target against HIV [8] [9] even though the highly active antiretroviral treatment (HAART) has allowed people living with HIV to live longer with a better quality of life. HAART supports or reinstates the immune system function. However, toxicity and resistance arise from HAART use [10] . Therefore, new antiviral therapy is needed since no cure or vaccine is available against HIV.
Furthermore, recent studies have shown a potential approach for controlling HIV-1 replication under combination antiretroviral therapy (cART)-free conditions to reach a functional cure by using a self-activating lentiviral vector that will deliver human A3G D128k gene target cells [11] . In the study of An et al., APOBEC3G polymorphism, such as rs8177832 (H186R), was found to be associated with HIV-1 subtype B [12] and C pathogenesis in some ethnic groups [12] [13] . Yet, this association was not ascertained in other populations. Additionally, subtypes A, B, CRF01_AE, and CRF_02AG of HIV viral infectivity factors’ have shown non-significant but somewhat differential levels of APOBEC3G based on infectivity profiles, whereas HIV subtype C was highly significant [14] [15] . APOBEC3G genetic polymorphism data is available for most world populations. However, a small coverage of African people possesses the most critical genetic variation due to many diverse genes and alleles [16] [17] . Therefore, findings from other populations may not always be generalized to Africans. Furthermore, in projects such as the 1000 genomes (2010) Projects and the Haplotype Map (HapMap) Project, the analysis of genetic markers genotyped are carried out on a small number of selected African populations [18] [19] [20] . Granting that these projects yield essential data on their whole human genetic diversity, they are narrowed down to the description of the African population [21] . This systematic review aimed to synthesize the current evidence on the effect of APOBEC3G polymorphisms and expression on HIV infection disease progression on the African continent. It will contribute to researching antiretroviral strategies to reach an HIV-1 functional cure while considering host and viral genetic diversity.
2. Methods
2.1. Search Method
The Preferred Reporting Items for Systematic Reviews and Meta-analyzes (PRISMA) were used to conduct this systematic review [22] . We included articles on the effect of APOBEC3G polymorphisms and expression on HIV infection disease progression on the African continent. We searched Embase, Google Scholar, PubMed, and Web of Science for relevant publications in French or English. The databases were searched on May 19, 2023. We used the following plain text and Mesh terms: APOBEC3G, HIV, and Africa in the respective entry terms or “apobec3* AND HIV* AND (Africa OR Sub-Saharan OR Algeria OR Angola OR Benin OR Botswana OR “Burkina Faso” OR Burundi OR “Cabo Verde” OR Cameroon OR “Central African Republic” OR Chad OR Comoros OR “Democratic Republic Congo” OR “Republic Congo” OR “Côte d’Ivoire” OR Djibouti OR Egypt OR “Equatorial Guinea” OR Eritrea OR “Eswatini (formerly Swaziland)” OR Ethiopia OR Gabon OR Gambia OR Ghana OR Guinea OR “Guinea-Bissau” OR Kenya OR Lesotho OR Liberia OR Libya OR Madagascar OR Malawi OR Mali OR Mauritania OR Mauritius OR Morocco OR Mozambique OR Namibia OR Niger OR Nigeria OR Rwanda OR “Sao Tome and Principe” OR Senegal OR Seychelles OR “Sierra Leone” OR Somalia OR “South Africa” OR “South Sudan” OR Sudan OR Tanzania OR Togo OR Tunisia OR Uganda OR Zambia OR Zimbabwe). The PubMed search strategy can be found in Table I. For the Google Scholar advanced search, the terms “APOBEC3G, HIV” were searched with all the times present in all the articles’ titles. The search was done in French and English, and the concerned papers were published between January 2009 and May 2023. Additional references were extracted from citations from original documents.
2.2. Study Selection and Data Extraction
Two independent reviewers screened the papers for titles and abstracts and then for full text for inclusion. Disagreements were resolved through discussion. We included articles reporting on APOBEC3G concerning African people infected by HIV. Case reports and reviews were excluded because they did not provide comprehensive results for the first and an analysis of articles for the second. Additionally, we were interested in studies on human subjects; therefore, in vitro studies or/and non-human subjects were excluded.
Two independent reviewers extracted the data separately, and disagreements were analyzed by the third reviewer. The following data were collected: first author’s last name, year of publication, country of origin, the purpose of the study, the study design, the biomolecular or genetic method used, the number of individuals analyzed, APOBEC3G polymorphisms, haplotypes, APOBEC3G mRNA level, and HIV infection.
The full-text analysis was done by two independent reviewers, judging the inclusion and exclusion criteria for the articles to be included in the study. The consensus was used in case of disagreements, or the third reviewer’s opinion would be considered. A third reviewer analyzed variances.
The average of the minor allele frequency of H186R of APOBEC3G was obtained from aggregated data from the studies that were available using the STATA software (College Station, USA).
2.3. Assessment of Risk Bias
Three independent reviewers assessed the risk of bias of the data on biological variation from the included articles through specific validated instruments. Biological variation studies on APOBEC3G were evaluated using the Biological Variation Data Critical Appraisal Checklist (BIVAC) published by Aarsand et al., 2018 [23] .
The included articles were from cross-sectional studies and were evaluated for their research questions, aims, population description, selection, and statistical analyses.
2.4. Summary Measures
Due to the heterogeneity of the studies, only the data from cross-sectional studies on the minor allele (G) frequency of H186R were aggregated, and qualitative synthesis was made on APOBEC3G biological variations about HIV in Africa. The other polymorphisms of Apobec3G were uniquely studied in Burkina Faso, Zimbabwe, and South Africa, and they were all different and could not be aggregated.
3. Results
The included studies were published between 2008 and 2019. The initial research identified 422 articles, with 77 duplicates and 345 papers remaining. In the following step, titles and abstracts were assessed to include relevant articles describing the role, expression, genetic variants, and haplotypes of APOBEC3G about HIV in Africa. Of the 288 articles considered, 57 were selected for full-text analysis. Finally, fourteen articles and a conference abstract on APOBEC3G biological variations about HIV in Africa were included in this systematic review. Figure 1 shows the PRISMA flowchart of the articles’ selection. Twelve of the selected studies were cross-sectional, and the three were longitudinal.
3.1. Critical Appraisal of the Included Studies
Following the risk of bias assessment, studies were scored based on high (A),
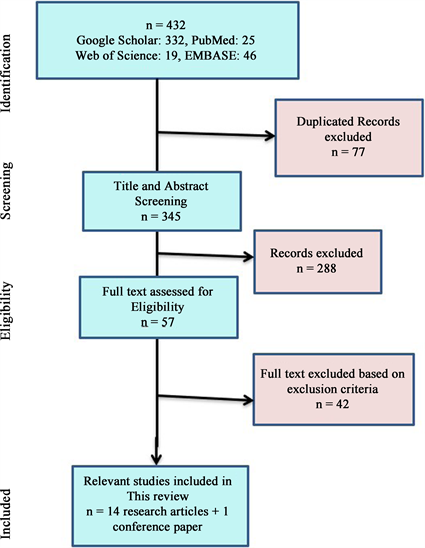
Figure 1: PRISMA Flow chart
moderate (B), or low (C) risk of bias due to methodological limitations (Supplementary file 1). In several of the included studies, estimates of analytical variation based on replicate analysis and estimates need to be presented, limiting the validity of their findings [24] - [31] . In addition, several studies are at risk of sampling bias. In Compaore et al. [24] [25] , groups were chosen by convenience sampling, while in Mhandire et al. [29] , patients were selected from a pediatric population, which limits the generalizability of these results. Furthermore, the data confidence limits for the study by Matume et al. [28] needed to be specified, making it hard to appreciate the different values reported.
3.2. Study Characteristics
The fifteen studies included were all from Sub-Saharan Africa (Burkina Faso, Central African Republic, Democratic Republic of Congo, Kenya, South Africa, Tanzania, Uganda, and Zimbabwe). There were twelve (12) cross-sectional and three (3) longitudinal studies. The sample size ranged from 24 to 708 participants.
3.3. APOBEC3G Polymorphism H186R Minor Allele Frequency and Its Role in HIV Infection
Seven studies were carried out on APOBE3G polymorphisms, and they were cross-sectional [13] [24] [25] [28] [29] [31] [32] . The average of the minor allele frequency of H186R of APOBEC3G available for the studies included in this study was 0.29 with a 95% CI (0.172; 0.401), with the lowest frequency reported by Uganda at 0.108 [32] and the highest recorded from Burkina Faso at 0.47 [24] [25] (Figure 2).
Figure 3: APOBEC3G Polymorphisms present in South Africa.
Legend
Blue: Minor allele G, Green: reference allele A
The preliminary data presented by Walsh et al. [32] reported an H186R minor allele frequency of 0.108 in Uganda and 0.148 in Tanzania (Figure 2).
Reddy et al. and Mhandire et al. have investigated the mutation H186R of APOBEC3G and have found that the mutation was not associated with HIV infection [13] [29] . Nevertheless, they found an association between the polymorphism H186R with high viral load and a low CD4 T cell count. Their finding confirmed the one of An et al. on an African American population. However, Compaore et al. have found that H186R was associated with HIV-1 infection. The minor allele of this polymorphism (G) seemed to be protective against HIV-1 infection, and the data was statistically significant OR = 0.8 (95% CI 0.641, p = 0.035) [24] .
![]() Legend: Blue: Minor allele G, Green: reference allele A.
Legend: Blue: Minor allele G, Green: reference allele A.
Figure 2. APOBEC3G Polymorphisms present in South Africa.
![]()
Table 1. H186R minor allele frequency in some African countries.
Footnotes: PCR-RFLP: Polymerase Chain Reaction-Restriction Fragment Length Polymorphism, NGS: Next Generation Sequencing, gDNA: genomic DNA.
Zhao et al. also reported an association between the polymorphism H186R and the HIV-1 status among ethnic groups of Central Africa (Biaka and Mbuti Pygmies) [31] . Indeed, unlike Compaore et al., Zhao et al. found that the reference allele (A) would be protective against HIV-1 among the Biaka ethnic group (OR = 0.86), and it seemed to be statistically significant [31] .
We have yet to find data from North Africa regarding the association between the polymorphism H186R and HIV-1 infection.
3.4. Presence of Additional APOBEC3G Variations and Their Importance
Four studies reported on other APOBEC3G variations other than the H186R (reference). Mhandire et al. genotyped five single nucleotide polymorphisms (SNP) in Zimbabwe, while Compaore et al., in Burkina Faso, genotyped 2 SNPs (Table 2) [24] [25] [29] . From South Africa, Reddy et al. identified 24 single nucleotide polymorphisms (SNP) by re-sequencing and genotyping in their cohort; of these SNPs, eight were newly identified, while the remaining 16 were already described (Table 2) [13] . Additionally, Matume et al., identified eight single nucleotide polymorphisms previously described in Northern South Africa (Table 2) [28] .
There was no association between the other than H186R polymorphisms studied and HIV-1 infection in the South African and the Zimbabwean population. Meanwhile, Compaore et al. found that the minor alleles of the polymorphisms rs6001417 and rs35228531, respectively, G and T, would be protective against HIV-1 infection, and the difference was statistically significant. We did not find studies on APOBEC3G polymorphisms other than H186R on central, eastern, and northern African populations. There was no association between the other than H186R polymorphisms studied and HIV-1 infection in the South African and the Zimbabwean population. Meanwhile, Compaore et al. found that the minor alleles of the polymorphisms rs6001417 and rs35228531, respectively, G and T, would be protective against HIV-1 infection, and the difference was statistically significant. We did not find studies on APOBEC3G polymorphisms other than H186R on central, eastern, and northern African populations.
3.5. APOBEC3G Haplotypes and HIV Status
Two studies reported on APOBEC3G haplotypes and HIV status. Mhandire et al. have investigated the presence of haplotype and linkage disequilibrium from their 6 SNPs of interest [29] . They found that when comparing the haplotypes with frequencies less than 0.03 between HIV-infected children and uninfected children, there was no association with the risk of HIV infection. Despite the latter, the APOBEC3G single nucleotide polymorphisms mainly were in strong linkage disequilibrium except for two pairs.
Regarding APOBEC3G haplotypes, Compaore et al. found that the 3 SNPs studied were in strong linkage disequilibrium [24] . Furthermore, the haplotype GGT would have a protective effect against HIV-1 infection, while two other haplotypes, GGC and CGC, seemed to increase the chances of HIV-1 infection from almost two (2) to five (5).
![]()
Table 2. Other APOBEC3G polymorphisms studied in Africa.
3.6. APOBEC3G Expression and HIV Infection
The level of APOBEC3G expression was evaluated regarding HIV infection in four (04) studies in this review. Research carried out by Ulenga et al. reported from a cohort of female sex workers in Senegal that the level of APOBEC3G mRNA was higher in individuals with a high viral set point compared to individuals with a low viral set point. They suggest that APOBEC3G protein would contribute to controlling viremia. Furthermore, APOBEC3G mRNA expression level was analyzed in a cohort of HIV-1 exposed seronegative individuals in Senegal. The authors reported a higher expression level of APOBEC3G in HIV-untreated individuals compared to HIV-infected individuals taking an antiretroviral treatment [30] . They also discovered that untreated HIV-1 infected subjects show a higher APOBEC3G mRNA level.
Similarly, Compaore et al., found that APOBEC3G expression was elevated in individuals naïve to HIV treatment compared to HIV-negative controls. Individuals with a high CD4 T cell count also had higher APOBEC3G expression.
Furthermore, HIV-infected individuals’ carriers of the minor allele G had a higher level of APOBEC3G than the controls carrying the same allele. Nevertheless, Reddy et al., found that APOBEC3G mRNA levels were higher in HIV-negative individuals compared to HIV seropositive individuals and that APOBEC3G transcription is speedily downregulated after HIV-1 infection. Ondoa et al. in Rwanda suggested that APOBEC3G expressed in a heterogeneous population of blood and mucosal cells might poorly correlate with viral loads because only a fraction of the cells analyzed represent targets and sources of HIV [33] .
3.7. Clinical Importance of APOBEC3G Hypermutation and Haplotypes
Four studies reported the clinical importance of APOBEC3 G-caused hypermutation and its haplotypes [26] [27] [31] [34] [35] . The CD4 T cell count was found to be correlated positively with hypermutated provirus in the study carried out in Kenya by Land et al. Subjects with HIV hypermutated (elevated adenine content) had higher CD4 T cell counts when considering CD4 T cell count as a surrogate marker of disease progression, and the difference was significant with p = 0.009 [27] . The authors also suggest that an increase in APOBEC3-type hypermutation was not due to Vif polymorphisms.
In Senegal, Ulenga et al., have found that APOBEC3G significantly contributes to the generation of G-to-A mutations in HIV-infected individuals. However, there was no correlation between the G-to-A mutations and the plasma viral load. The authors suggest that APOBEC3G might restrict HIV viral growth in a G-to-A mutation-independent manner [35] .
In South Africa, Reddy et al., have shown that A3G WT (APOBEC3G wild type) and A3G-186R (APPOBEC3G-186R mutant) are counteracted at the same level by Vif irrespective of the patients’ Vif variant [34] . Nevertheless, a lower antiviral activity was shown by the A3G-H186R variant. The latter could justify that the patients expressing A3G H186/H186R reported higher plasma viral loads and accelerated disease progression. Furthermore, Lee et al., in a comprehensive evaluation of early HIV-1 DNA genome landscapes during the first year of infection in South Africa, have found that APOBEC3G activity, as measured by the presence of hypermutated viral genomes, was not observed in the earlier phase of disease but increased over time [26] .
4. Discussion
This study reviewed the literature to study the role of APOBEC3G polymorphisms, haplotypes, and expression in HIV infection and disease progression on the African continent. It is worth noting that the data on APOBEC3G concerning HIV is scarce across Africa, especially in Sub-Saharan Africa. However, it bears the highest toll of new infections and AIDS-related deaths [36] . Overall, 32 single nucleotide polymorphisms of APOBEC3G were identified in Africa regarding studies related to HIV [13] [24] [28] [29] [31] .
The data gathered show that H186R minor allele frequencies found among Africans overall are higher than those reported by Caucasians (2.7%) and Asians (7.8%) [12] . The latter data seem to reflect that of HIV case distribution worldwide. Likewise, Skelton et al., reported a BST-2 variants differential distribution, which mirrors HIV infection prevalence in Africa [37] .
The APOBEC3G polymorphism H186R frequency varies from 0.108 in Uganda to 0.47 in Burkina. Meanwhile, An et al. found that the H186R minor allele frequency was about 37% among African Americans [12] . The frequency of H186R reported seems to decrease from western Africa to southern Africa, crossing central Africa (Democratic Republic of Congo). APOBEC3G single nucleotide polymorphism H186R lowest minor allele frequency is reported from Eastern Africa (Tanzania and Uganda) [24] [31] [32] . These disparities follow Campbell et al.’s map of African language family distributions [38] . The frequency of the minor frequency allele of H186R is low in areas where the Nilo-Saharan language is predominantly spoken with a mix of Niger-Kordofanian. Furthermore, H186R minor allele frequency is decreasing following the South migration and the language of western Bantu [16] [38] [39] . Population migrations during centuries could explain the differences in H186R MAF [38] . APOBEC3 variations regarding evolutionary origins or the diversification of the APOBEC3 enzyme principles are yet to be discovered [40] .
Studies on HIV subtype B infected African Americans reported that there was no association between the APOBEC3G mutation H186R and HIV infection [12] . Similarly, Reddy et al. on the South African population and Mhandire et al., in Zimbabwe came to the same conclusion on HIV subtype C infected individuals [13] [29] [41] . However, Compaore et al., found that the minor allele of H186R was protective against HIV infection in an HIV-circulated recombinant forms CRF06_cpx and CRF02_AG context. Meanwhile, in a population of the Central African Republic with a high degree heterogeneity of HIV-1, the reference allele of APOBEC3G polymorphism H186R was protective against HIV infection [31] [42] . There was no data from Eastern Africa regarding the association between the mutation H186R and HIV-1 status. Host and viral genetic variation can account for these discrepancies.
Except for Compaore et al., none of the studies found an association between polymorphisms of APOBEC3G other than H186R and HIV infection [24] [25] . Indeed, the other two polymorphisms (rs6001417 and rs35228531) are in high linkage disequilibrium with the polymorphism H186R in the study population from Burkina Faso. The ladder could explain that rs6001417 and rs35228531 minor alleles seem protective against HIV-1 infection in Burkina Faso. There was no information from studies on other polymorphisms than H186R in Northern, Central, and Eastern Africa.
Mhandire et al., have investigated the presence of haplotype and linkage disequilibrium from their 5 SNPs of interest. They found that the six haplotypes with frequencies less than 0.03 observed in their population were not associated with HIV infection [29] . Additionally, the 5 SNPs studied were located on the gene at about 7KB, from rs2294367 to rs5757463, and overall, they were in linkage disequilibrium except for two. Meanwhile, Compaore et al., found strong linkage disequilibrium between the 3 SNPs they studied. The minor alleles GGT haplotype was protective against HIV infection, and some other haplotypes were increasing the probability of being infected by HIV [24] [25] . The difference between these two studies could be due to the fact that the SNPs studied by Compaore et al., are closely situated on the APOBEC3G gene, about 6 kb between rs6001417 and rs35228531, the two extremes with rs35228531 being an extra-genetic mutation close to the 3’end [13] . Populations’ genetics could also account for these discrepancies [21] . Additionally, selection mechanisms, or a bottleneck effect, might explain why some APOBEC3 haplotypes are primarily present in some Saharan populations and others outside the continent. APOBEC3 is located on chromosome 22 and possesses haplotype blocks that span over 2 or more APOBEC3 genes. There is a lot of splicing events that occur between APOBEC3 genes adjacent exons, since the genes are so close. Nonetheless, the roles in viral hypermutation of inhibition by APOBEC3G genomics and transcriptomics interdependence are still unknown [40] .
Meanwhile, studies in West Africa report that APOBEC3G mRNA levels are high among HIV-infected untreated individuals compared to HIV-infected individuals receiving treatment [30] [43] [44] , Reddy et al., in Southern Africa found that APOBEC3G mRNA levels are lower in HIV individuals compared to HIV seronegative persons [13] . The latter results were similar to that of previous studies showing that APOBEC3G levels were higher in seronegative individuals compared to HIV seropositive individuals [45] . While a study in Rwanda found that APOBEC3G expressed in a heterogeneous population of blood and mucosal cells might poorly correlate with viral loads [33] . These differences could be due to population and viral genetics.
In a study carried out by Land et al., in the context of HIV-1 subtype A, as it is the most represented HIV subtype in Kenya, the CD4 T cell count was found to be correlated positively with hypermutated provirus [27] [46] . Subjects with hypermutation (elevated adenine content) had higher CD4 T cell counts when considering CD4 T cell count as a surrogate marker of disease progression, and the difference was significant. The authors also suggest that an increase in APOBEC3-type hypermutation was not due to Vif polymorphisms. This study on a Kenyan population is paving as it allows an understanding of the clinical importance of APOBEC3G hypermutation regarding HIV Vif variants. It confirms the argument that APOBEC3-mediated hypermutation has an antiviral role [47] [48] . However, in South Africa, Reddy et al., have shown that A3G WT and A3G-186R are counteracted at the same level by Vif irrespective of the patient’s Vif variant [34] . Nevertheless, a lower antiviral activity was shown by the A3G-H186R variant. The latter could justify that the patients expressing A3G H186/H186R reported higher plasma viral loads and accelerated disease progression. This finding supports the previous study of Reddy et al., that APOBEC3G polymorphisms may considerably affect HIV-1 pathogenesis [13] .
Meanwhile, Ulenga et al., suggest that APOBEC3G might restrict HIV viral growth in a G-to-A-independent manner [35] . Nevertheless, these conclusions cannot be extrapolated to the whole African population as the latter studies were carried out on a small sample size and were yet to be carried elsewhere on the African continent to our knowledge. Furthermore, it was shown that African populations are genetically diverse [49] .
5. Conclusion
This systematic review shows that it is a potential target for an anti-HIV strategy as it is a potent antiviral factor despite HIV-1’s Vif action. However, there is a need to increase the research on APOBEC3G from its variants to its hypermutation on the continent with an essential variety of HIV-1 subtypes to impact the research on APOBEC3G-based anti-HIV strategies.
Statement of Ethics
Ethics approval was not required, since the work is a review.
Acknowledgements
We want to thank all our colleagues for their critical manuscript reviews.
Funding Sources
No funding was received for this work.
Authors Contributions
T R C: Screen papers Conceptualization (lead); writing—original draft (lead); Formal analysis (lead); screened the papers on titles and abstracts, and then on full text for inclusion; extracted the data separately; full-text analysis judging the inclusion and exclusion criteria for the articles to be included in the study; assessed the risk of bias of the data on biological variation from the included articles through the specific validated instrument.
A K O: Formal analysis (supporting); writing—review and editing (equal), screened the papers on titles and abstracts, and then on full text for inclusion; extracted the data separately; complete text analysis judging the inclusion and exclusion criteria for the articles to be included in the study; assessed the risk of bias of the data on biological variation from the included articles through the specific validated instrument.
A B: Methodology (lead); writing—review and editing (equal); full-text analysis judging the inclusion and exclusion criteria for the articles to be included in the study and analyzing disagreements); assessed the risk of bias of the data on biological variation from the included articles through specific validated instruments.
L T: Software (lead); writing—review and editing (equal); assessed the risk of bias of the data on biological variation from the included articles through specific validated instruments.
A A Z and H G O: Conceptualization (supporting); Writing—original draft (funding); Writing—review and editing (equal).
S K and J S: Supervision (Lead); Writing—original draft (supporting).
All authors give final approval of the manuscript to be submitted.
Data Availability Statement
All data generated or analyzed are included in this article. Further inquiries can be directed to the corresponding author.