Archaeogeneticists on the Origin of the Indo-Europeans, with Particular Emphasis on the Pre-Slavs as Their Oldest Ancestors ()
1. Introduction
Population genetics, similar to paleolinguistics (Alinei, Kortlandt and others), supports the thesis that the Slavs settled in Central Europe from at least the Bronze Age. There are also voices that it is the Slavs who are genetically and linguistically the most similar to the oldest Pra-Indo-Europeans. This view, however, hardly makes it into the scientific discussion, due to the resistance of Western scholars related to historical prejudices against the Slavs, as well as the implementation of a specific historical policy and false scientific paradigms adopted on its basis, such as the one about the arrival of allegedly primitive Slavic tribes from the east to Central Europe in the 6th century CE, where they were supposed to occupy the abandoned lands of the “higher civilizationally developed” Germans (Kossinna).
This servility of scholars, sometimes wrongly called “scientific patriotism”, prevents a sober approach to the subject of the ethnogenesis of European peoples, who until now have been associated for political reasons, mainly with the participation and achievements of the Italo-Celts and Germans, excluding the Slavs, who ancient times were located somewhere far away in Asia.
Today, in the face of the results of archaeogenetic research, such erroneous and irresponsible hypotheses, conditioned by nationalist premises from the period of Nazi domination in Nazi Germany, Austria and Italy, but also in some English, French and American intellectual circles, must be revised and put aside to science.
After the initial analysis of the data, I began to see the image that the cradle of the Slavic lands are the territory between the Oder and the Dnieper (Polish: Odrodnieprze), and the range of occurrence of the carriers of the R1a1 haplogroup practically coincides with the current distribution of Slavic nations (compare Map 1 and Map 2).
It is this haplogroup that is also typical of the Indo-Aryans. It can therefore be concluded that instead of talking about “Indo-Germans”, as was customary in the 19th-century science and which, in a way, persists in it until today, we should
![]()
Map 1. Haplogroup R1a1 distribution in Europe (Wikipedia).
![]()
Map 2. Slavic countries in Europe (the map almost covers the area of dominance of hg R1a1).
use the term “Indo-Ario-Slavs”, and bearing in mind the possible route of migration of representatives of hg R1a1 from west (central Europe) to the east (Persia and India), an even more accurate term would be “Wendo-Ario-Indians”. Of course, assuming that the Wends are Proto-Slavs.
Of course, many scientists believe that this is a coincidence, and the blood type does not prove ethnicity. But if we ask them what the distinguishing features of ethnicity are, they either avoid the answer or don’t know it. Increasingly, it is seen that in scientific discussions on this subject, not only blood ties, but also language and culture are rejected.
The conclusions drawn boil down to laconic statements that on the basis of genes, bones or ceramics alone, little can be said about the origin of given communities, or that ancient peoples did not know what an ethnos is, so they cannot be attributed a sense of ethnicity, at least ancestral. If, on the other hand, the subject of ethnically-based nation-building processes is discussed, scholars neatly cut the subject short, claiming that the concept of “nation” is a new construction to which ethnogenetic considerations do not apply.
In this neat, yet calculated way, archaeogenetic issues and the whole topic of the origin of peoples are pushed to the margins of science, often accusing the authors who deal with it with nationalistic or even racist tendencies, which is a convenient tool for depreciating their work.
2. An Overview of the Most Important Views of Archaeogeneticists on the Origin of the Indo-Europeans
2.1. The Beginnings of Population Genetics
One of the precursors of population genetics was Luca L. Cavalli Sforza (Cavalli Sforza & Feldman, 1981; Cavalli Sforza, 1994) , a professor at Stanford University, who initiated research on the coevolution of genes and culture in the 1980s. He indicated a close relationship between male haplogroups (Y-DNA) and languages. His research showed that Slavic and Indo-Iranian languages are usually used by the holders of hg R1a.
In 2000, a group of scholars led by Ornella Semino (2000) proposed a post-glacial (Holocene) spread of haplogroup R1a1a from the north of the Black Sea during the Late Glacial Maximum, which was then compounded by the expansion of the barrow culture into Europe and east.
In November 2009, the journal “Nature” published an article about the discovery of a new version of the R1a1 haplogroup—R1a1a7. Researchers examined the time and place of origin of various mutations of the male haplogroup R1a, which is possessed by almost 57 percent of the population contemporary Poles. An international team of scientists, including Peter A. Underhill et al (2010) from Stanford University, estimates that it has existed in Poland for about 10,700 years, i.e. since the melting of the glacier.
In this study, American geneticists used the theory of coalescence (separation), which allows to determine how gene variants taken from a population could have come from a common ancestor. In the cited work of the Underhill team, it was written that at least 7000 years ago (ybp—years before present), the ancestors of more than half of today’s Poles lived in Poland, Slovakia and... in Crete. In turn, the mutation of this haplogroup, the most characteristic for Poles, with the symbol R1a1a7, was placed at the latest 6.6 thousand ybp in the Polish lands, linking it with the Świder culture (Bogdanowicz, 2018) .
One of the markers of hg R1a-M458 is considered to be a Polish subclade, as it occurs in the greatest intensity among the population of Poland (Map 3). Underhill writes that marker M458 has a significant frequency in Europe, exceeding 30% in its main area in Eastern Europe and covering up to 70% of all M17 chromosomes present there. M458’s diversity and frequency profiles suggest an Early Holocene origin [ca. 10 - 11 thousand ybp] and subsequent expansion possibly related to many prehistoric cultural events in the region. Its original frequency and diversity distribution correlates well with some of the major central and eastern European basins where sedentary agriculture was established before its spread further east. Importantly, the apparent absence of M458 chromosomes outside of Europe argues against a significant flow of patrilineal genes from Eastern Europe to Asia, including India, from at least the mid-Holocene (Underhill et al., 2010) .
It was also noted that “the subhaplogroup R1a1a7-M458 has the highest frequency among the Slavic and Finno-Ugric peoples”. In this case, the term “Finno-Ugric” is limited to one nation from this language group—the Hungarians, who were to lose their Slavic language in favor of Magyar only in the Middle Ages and are an exception to the rule of Cavalli Sforza (Bogdanowicz, 2018) .
Peter Underhill et al. (2010) state that our phylogeographic data lead us to con
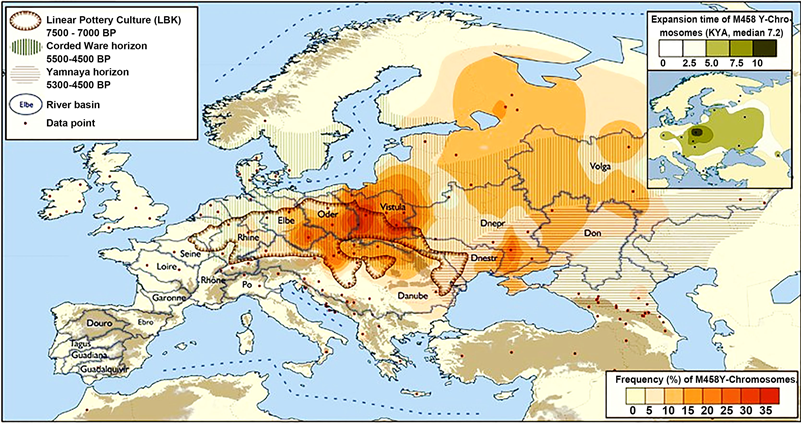
Map. 3 Distribution of haplogroup R1a-M458 in Europe with an outline of archaeological culture areas (Underhill et al., 2009, 2010) .
clude that the initial episodes of R1a-M420 diversification occurred near Iran and eastern Turkey, and we estimate that diversification below M417/Page7 occurred around 5800 ybp. This suggests the possibility that the R1a lineages accompanied demic expansions initiated in the Copper, Bronze and Iron Ages, partially replacing the previous layers of the Y chromosome, an interpretation consistent with the, albeit limited, ancient DNA evidence.
Further in the study by the Underhill team, we read that although R1a occurs as the most common Y-chromosome haplogroup among populations representing a wide variety of language groups, such as Slavic, Indo-Iranian, Dravidian, Turkic and Finno-Ugric, many authors have been particularly interested in the link between R1a and the Indo-European language family. For example, R1a frequency patterns have been discussed in the context of a purported link between Indo-European-speaking shepherds and archaeological evidence for the distribution of the Kurgan culture in the Pontic steppe. A more precise interpretation of the underlying prehistoric and historical episodes of R1a chromosomes across this wide range of Eurasian geography remains largely unknown due to insufficient information on the phylogenetic subdivisions within the R1a haplogroup. We address this shortcoming by analyzing over 11,000 DNA samples from across Eurasia, including over 2000 from the R1a haplogroup, to ascertain the phylogenetic information of newly discovered R1a-related SNPs. We are also studying the STR diversity of related R1a subclades to better understand the demographic history and prehistoric cultural links of one of the most widespread and frequent Y-chromosome haplogroups in the world with post-Last Glacial Maximum origins (Underhill et al., 2010) .
The subclade R1a1a (R-M17 or R-M198) is the subclade most commonly associated with representatives of Indo-European cultures. It just so happens that Poles have one of the biggest dominants of this haplogroup in Europe (as you can see on the map: Distribution of the R1a1 haplogroup in Europe, posted above). And this, in turn, means that among the European peoples, we are genetically the closest to the Proto-Indo-Europeans. Therefore, Indo-European studies should be in our area of interest, especially when it comes to including Slavic roots in the etymologies of words in European languages (abolishing the unwritten rule of ignoring Slavic roots based on the false paradigm of the late arrival of Slavs to Central Europe), but also determining the origin of peoples, especially based on kinship within haplogroup R1a1.
In my opinion, Underhill et al. (2010) have probably most accurately located the original homeland of the R1a representatives in the area from western Anatolia, ancient Syria, Colchis and Armenia and the South Caucasus to the area of present-day eastern Iran (formerly: Sumer, Akkad, Mesopotamia, Babylonia). This partly coincides with the hypothetical birth area of R1a adopted by Pamjav et al. (2012) , who extended this territory to the lands north of the Caucasus, occupied in antiquity by Scythian tribes (Map 4).
The range given by Underhill is the area inhabited by the Aryans, but let’s remember that the time of the ancient civilizations of the Near East is at most the 4th millennium BC, and the period of the splitting of R into R1a and R1b dates back 22,000 years. Therefore, by tracking the dating of individual R1a samples, it is possible to determine the routes and periods of migration of representatives of this haplogroup. Horváth (2016) assumes that they most likely entered the Balkans through Anatolia. Semenov and Bulat (2016) assume that their route, bypassed the Caspian Sea on the right side and continued north-west. In turn, Underhill et al. (2010) locate the oldest links of expansion in the area between the
Vistula and the Rhine, in the Caucasus and in the Indus Valley. On the other hand the largest contemporary concentrations of R1a Underhill (2014/2015) are located: 1) from the Vistula River through the entire East European Plain, 2) the southern slopes of the Himalayas, 3) the Karakoram and Tienshan mountain valleys, 4) Altai.
In Europe, the R1a1 haplogroup (Y-DNA) is most saturated among the Slavic nations: Lusatian Serbs (63%), Poles (55%), Belarusians (50%), Russians (46%) and Ukrainians (43%). Its significant share is also present among Finno-Ugric peoples, mainly Estonians (33%) and Hungarians (25%). Also in the Baltic countries, a high frequency of the R1a haplogroup was found: Lithuanians (34% - 45%), Latvians (40%). A certain percentage of R1a is also found in Scandinavians: Norwegians (25% - 30%), Swedes (15% - 20%), Danes (10% - 15%) and Germans (15% - 20%). However, outside Europe, R1a is most common among the inhabitants of North India (48% - 73%) and Central Asia: Iranian-speaking Ishkashim (68%), Tajiks (30%) and Pashtuns (40% - 45%), Turanian peoples: Kyrgyz (63%) and Altaians (38% - 53%) (Pericić et al., 2005) .
Underhill’s (2010) team determined that both subgroups R1a1a-M17 and R1a1a1g-M458 have been present in each country since the following times (in years):
● Poland—11,300 and 10,700;
● Slovakia—11,200 and 8300;
● Czech Republic—5700;
● Germany—9900 and 7500,
● European Russia—8700 and 8000;
● Ukraine—7400 and 4700;
● India—14,000;
● Pakistan—15,000.
Data on the dating of the R1a1a-M17 and R1a1a1g-M458 populations in individual countries seem to be overestimated, but they are still a great breakthrough in the dispute about the origin of the Indo-European peoples, giving strong arguments also to the supporters of the autochthonism of the Slavs.
Even with this certain overestimation of the dates of occurrence of these R1a1a subgroups, one cannot understand the passivity on this issue of many academics who seem to have entrenched themselves in their positions. From where they stubbornly try to convince others of their false reasons, negatively verified by modern science, and archaeogenetics and paleolinguistics in particular, which agreed with those archaeologists, linguists, and anthropologists who have long been preaching the theory of indigenous Slavs.
2.2. Discovery of the “Polish” Mutation R1a—L260
Shortly after the description of hg R1a1 by Underhill’s team, another mutation was detected in this “Slavic” subgroup (R1a1a1b1a1/R1a1a7-M458), designated L260 (R1a1a1b1a1a /R1a1a1g2), which was named P—Polish (Figure 1). It occurs
![]()
Figure 1. The cleavage path of hg R to form R1a1a1b1a1a, in other words the L260/S222 clade, called “Polish” (Eupedia) (Hay, 2017) .
from about 800 - 600 BC only in Poland, the Czech Republic and Slovakia, which proves the long-term stability of life in a given area by the same genetic human population. This subclade, however, does not exist in Eastern Europe, so it could not have been brought west by migration from the east. L260 is already among the Iron Age Przeworsk culture skeletons in Poland (as well as among 17% of modern Polish men). This discovery further confirmed scholars that the origins of the Western Slavs, including the ancestors of Poles, in Central Europe date back to prehistoric times.
Thus, we can see that from R1a1a1b1a1-M458 (from c. 2800 BC) R1a1a1b1a1a-L260 separated (from c. 800 BC), which is specific only to Poland, the Czech Republic and Slovakia, which is why this clade is called “Polish” (Figure 2).
From it developed the subclade R1a1a1b1a1a1a3/YP414 (L260 > YP256 > YP254 > YP414), which is even more considered a Polish subclade of the R1a-M458 haplogroup. It occurs in every tenth Pole. Also in terms of numbers, Poles account for almost half of the share. YP414 has a population of 5.3 million and is the second largest for M458. It was created quite late, around 300 CE, but it was undoubtedly successful. It probably originates from Poland or, more broadly, from the area to the Elbe and the middle Dnieper, and from there it migrated in different directions.
An interesting visual diagram of the hg M420 branching with a visible sublayer L260 (Map 5) was also presented by Igor Rozhanskii (2017) . According to this Ukrainian geneticist working in Japanese Tsukuba, the L260 subclade, called “West Slavic”, is related to the Central European genus (CTS11962), but has a more compact range and a smaller population—an estimated 11 - 12 million people. Its haplotypes are well recognized in the 17-marker format, which is why it manages to get estimates for the Balkans. In this region, however, its share among Croats only exceeds the threshold of 1%, and the maximum value was
![]()
Figure 2. The occurrence of R1a-YP414 in the population of some countries, among which Poland predominates (Vayda) .
again shown by the Lusatians—about 1/3 (41 out of 123), i.e. 33%.
The haplotypes of the West Slavs of the R1a1 (L260) haplogroup are poorly represented among other peoples of Europe (at most—7% in Germany), perhaps meaning the descendants of the West Slavs. Accordingly, the range of this branch is the Czech Republic, Slovakia, Poland, Ukrainian Transcarpathia with separate inclusions in the surrounding regions.
It is estimated that about 20 million people belong to the Central European genus CTS11962, which is comparable to the male population of countries such as Poland or Canada. In almost all haplomap regions east of the Rhine, its share exceeds the threshold of 1%, reaching a maximum of 15% in western Poland (Map 6). The absolute record belongs to Lusatia, a small Slavic people in the GDR, with a share of 1/4 (29 out of 123). The last estimate was made from a field trial in a 17-marker format where the Central European branch is well separated from the rest when calculating the tree. This feature made it possible to supplement the haplomap area with Western Europe and the Balkan countries. According to YFull, there are 6 main daughter branches in the CTS11962 branch, most of which are in the western part of the range. In the east, including among Russians, the YP417 branch is mainly found so far (Rozhanskii, 2017) .
2.3. In Search of the Genetic Pattern of “Central Europeans” and Their Original Habitats
In the article from 2009, I. Rozhanskii and A. Klyosov (2009) placed the common ancestors of the carriers of the R1a1 haplogroups in Central Europe and Scandinavia around 4500 ybp, for three base haplotypes of hg R1a1 (DYS464e, DYS464f and DYS464g). It is difficult to say whether it was Western Europe, Central Europe or the Russian Plain, but the fact is that 4600 ybp R1a1 carriers were in Germania according to these dates.
They also try to estimate, based on the available data, where the first “Central
European” lived 2400 - 3000 ybp. Since this branch is quite reliably divided into two related sub-branches with one common ancestor, then, according to Ockham’s razor, one should look for the ancestral homeland where these sub-branches are most geographically intermixed. Map 4 shows a fairly large area from Bavaria in the west to Transcarpathia in the east, covering the entire Czech Republic, Silesia, Slovakia, partly Hungary and Romania.
Rozhanskii and Klyosov explain that the 7 mutations on the first 25 markers define a common ancestor for both populations (West Slavic ZS and Central European SE) R1a1 around 4900 ybp. This is the time of the beginning of active migrations of the R1a1 haplogroup in Europe and the life of the common ancestor of R1a1 on the Russian Plain. It seems that the fate of the descendants of the Eastern Proto-Slavs was more successful in terms of survival than their brothers in Europe. The group that went east, to the Russian Plain, maintained a direct genealogical line. However, those who remained in the center of Europe and in the western foothills of the Carpathians, preserved only fragments of genealogical lines—more precisely, one recent branch that survived in the middle of the 1st millennium BC.
Despite the significant geographical distance, the East Slavs, Germans and Poles are randomly mixed in sub-branches, which clearly indicates recent migrations of carriers of these haplotypes. It is logical to assume that such migrations were one of the episodes of the Migration of Peoples of the 4th-8th centuries. Since it seems that carriers of Central European haplotypes moved from west to east, contrary to the general flow in this period, confirmation of such migrations in specific years should be sought in historiography and archaeology.
Some of the Slavs, among whom there were carriers of local Central European haplotypes, left their homes and moved to places where peoples related to them linguistically and culturally lived. One group settled on the Dnieper, others went to Poland and Silesia, and some during numerous wars were captured by the Franks and, if not sold as slaves, were allowed to settle in lands depopulated after long wars with the Saxons. Apparently, this is how this branch spread among the Germans of Lower Saxony and their descendants who settled in Pomerania and East Prussia. Although direct evidence of a mass migration of Slavs from Pannonia and Transylvania has not yet been found, it can be indirectly assumed that the territory of the Avar Khaganate is currently inhabited mainly by Hungarians and Romanians. Of course, the outflow of the Slavic population after the defeat of the Avars was so large that relatively small groups of Ugro- and Roman-speaking settlers could easily assimilate those who remained.
Russian scholars believe that analyzes of the Central European subclade indicate a migration of the Polans from the west to the east, which is in accordance with Nestor’s account of the arrival of the Poles from the Danube to the Vistula, and from there also to the east. It is therefore logical to look for the “fields” to which the Glades get their name, somewhere in the plains of Pannonia (Rozhanskii & Klyosov, 2009) .
Rozhanskii is wrongly attributed with the saying “Поскреби русского—найдешь поляка” (“Scratch the Russian—you will find a Pole”), which is only the title of an article by Nestor Sheikin (2022) that appeared in the Russian press. However, he writes about the work of Oleg Balanovsky’s team, not Igor Rozhanskii. However, it seems to be a concise summary of research in the field of genetic archeology from Russia, pointing to the Vistula origin of the local Slavs.
Nestor Sheikin refers in his text to the work of Balanovsky et al. (2008) that contrary to Karamzin’s famous saying about the significant admixture of Tatar blood among the “pure Russians”, scientists have not been able to find traces of it. The results of a large-scale study showed that the largest ethnic group in Europe consists of two slightly mixed groups, and that Poles and Ukrainians are genetically closer to Russians from central Russia than their northern compatriots. One of the main findings of this pioneering work is the overriding importance of geographic, rather than linguistic, factors in explaining the observed Y-chromosome variation across ethnic groups in Europe.
The original territories of the formation of the Slavic peoples are also still hotly debated, but it is certain that the mass migration of Slavs from Central Europe to the east took place in the 7th-9th centuries. The resettlement took place in two directions—north-east and south-east. Apparently, the migration to the northeast was accompanied by a wide assimilation of local ethnic groups—Baltic and Finno-Ugric.
Thus, we are not talking about the movement of the Slavs from the Dnieper to the west in the 6th century, but about migration in the opposite direction, which was also mentioned by Rozhanskii (Map 7). Therefore, such a concept is not
![]()
Map 7. R1a1a migration according to Igor Rozhanskii. Northwestern (NW), Central Eurasian (CEA), Eastern Eurasian (EEA), Western Eurasian-2 (WEA-2), Northern Eurasian (NEA), Central European (CE), West Slavic (WS), Northern European (NE) and Younger Scandinavian (YS).
convenient for all Russians, as some of them prefer to identify themselves with the European Vikings (Varangians/Ruthenians) rather than with the Poles, with whom they have always had a disagreement. Unfortunately for them, data from fossil DNA research confirm Nestor’s version about the transition of some tribes of Lyakhs (Polyanians, Vyatichians, Radimichians) from the Vistula River to the east (Cross & Sherbowitz-Wetzor, 1953) .
The research of the Balanovsky team shows that in the prehistoric times of the existence of the Proto-Slavic peoples in Eastern Europe, there was no significant migration of people from Asian regions.
The results of the analyzes clearly show that “the genetic variability of the Y chromosome of the inhabitants of the central and southern regions of ancient Russia turned out to be not only almost identical to that of the »Slavic brothers«—Ukrainians and Belarusians, but also very similar in structure to the varieties of Poles” (Balanovsky et al., 2008) .
Scientists believe that this observation can be interpreted in two ways. Firstly, such proximity of the genetic structure may mean that the process of migration from the lands of Central and Eastern Europe to Western Russia was not accompanied by the assimilation of local peoples—at least those that had strong differences in the structure of the male genetic line. Secondly, it may mean that the Slavic tribes had already mastered these lands long before the mass migration of the ancestors of the Russians in the 7th-9th centuries. This point of view is consistent with the fact that the East and West Slavs show great similarity and smooth, regular changes in the structure of the male genetic line.
Balanovsky emphasizes that in all cases the genetically identified subpopulations do not extend beyond ethnic groups defined by language positions. This rule, however, has one very interesting exception: four large groups of Slavic peoples—Ukrainians, Poles and Russians, as well as Belarusians—show a large approximation both in the genetic structure of the male hereditary line and in language.
It would seem that such a situation should collide with the thesis that geographical factors have a greater impact on Y-chromosome variants than language ones, because the territory occupied by Poland, Ukraine and the central regions of Russia stretches almost from the center of Europe to its eastern border. The authors of the study, commenting on this fact, note that genetic differences apparently have much in common even with territorially distant ethnic groups, provided that their languages are similar.
The data presented in the results show that Poles show the lowest intra-ethnic variability among some nations in Europe selected for comparative analysis (Table 1). In other words, they are one of the least genetically diverse nations in Europe.
Balanovsky notes that that Russians, especially from the central-southern part of Russia, are closest to Poles, Ukrainians and Belarusians. Poles also show the lowest level of intra-ethnic variability in Europe, which proves their homogeneity.
Scholars themselves conclude that “the Y-chromosome pool of Russians in their historical settlement area is mainly a composite of their Proto-Slavic heritage, and especially in the north of Russia, extensive admixture with Finno-Ugric speakers”. Despite the widespread opinion about the strong Tatar-Mongol admixture in the blood of Russians, inherited by their ancestors at the time of the Tatar-Mongol invasion, the haplogroups of the Turkic and other Asian ethnic groups practically left no trace on the population of the modern north-western, central and southern regions. Instead, the genetic structure of the paternal lineage of the population of the European part of Russia shows a smooth change when moving from north to south, indicating two centers of the formation of ancient Rus. At the same time, the movement of the ancient Slavs to the northern territories was accompanied by the assimilation of local Finno-Ugric tribes, while in the southern territories individual Slavic tribes and nationalities may have existed long before the Slavic “great migration” (Balanovsky et al., 2008) .
In another of his works, Balanovsky (2012) placed a quite meaningful map (visible above), developed according to the “jigsaw principle” (Map 8), illustrating territories where the frequency of the haplogroup is greater than one-third of the gene pool (>35%). This area is shown in its own color for each haplogroup. Each geographical part of Europe is dominated by one haplogroup, rarely seen in other parts.
The data collected on Eupedia show that the highest density of the R1a1 population is in Poland—57%, and in the Lusatian Serbs this ratio is even higher—63.4%, as well as in Lesser Poland—64% and in Kashubia—over 60%. For comparison, in Leipzig, i.e. in the previously Slavic lands—27.1%), while in Mainz R1a—8.4%, and R1b—44.2% (Hay, 2017) .
Currently, researchers are arguing about the cradle of these peoples. As we know, both R1a1 and R1b1 derive from the very rare haplogroup R and its successor
R1. The dispute is between the supporters of the Altai and Iranian cradle of these peoples. Important information is the fact that haplogroup R developed 20,000 - 34,000 ybp, while R1 about 18,000 ybp. Haplogroups R1a1 and R1b1 are slightly younger than R1. As we know, this is the period of the last major glaciation. It is highly probable that the small number of people with R1 testifies to a sharp decline in the human population in that area. The large number of people with R1a1 and R1b1 seems to suggest emigration to warmer regions.
At this point, we can also refer to Anatole Klyosov (2019) , who claims that the Slavic people came to Europe from the Altai steppes through the Middle East (the Catal Hoyuk settlement) and the Balkans. The Russian scholar states that the Vinča culture was created by the Slavic people fleeing from the deluge that flooded the Black Sea coast 5600 years BC. It is generally believed that the Vinča culture gave rise to the Danubian cultures with the Corded Ware culture (Map 9) as their apogee in the 3rd millennium BC. Evidence for this thesis can be found in the confirmed cases of the genome with hg R1a1 of individuals attributed to this culture.
As for the haplogroups R1a1a1g and R1a1a1g2, until recently called R1a1a7, it should be said that these groups are very characteristic of the Western Slavic community (in southern Poland, over 30% of this subgroup alone). The formation of this haplogroup is tentatively dated to 2000 BC. The oldest varieties can be found in Serbia and Croatia (Lesser Poland is identified with the so-called White Croatia, and Lusatia with White Serbia). A very interesting fact about the
R1a1 genomes is the information that the variance of this gene in the Polish community is higher than for the entire European population. This means nothing more than that the most likely region where the rapid increase in this population has occurred is Poland. Scientists in the world are currently almost completely unanimous that the Slavic people populated the areas of western Russia from the territories of Poland. The invasions of the Scythians and Sarmatians constituted the backward emigration of the people who came to the steppes from Poland.
A new SNP, M458, announced by Underhill’s (2010) team, splits R1a-M17. According to him, R1a1a*, the part of R1a1a that is negative for M458, is widespread in Eurasia. Underhill reports that the highest coalescence time (age) for R1a1a7 among Poles is 10,700 years. Many of the modern M458 carriers in Poland come from two extension populations that are much more recent, as each type is much less diverse (lower ASD) than the total. Since both are large juvenile types, the simplest explanation is that each has recently grown out of a small founding population. On the other hand, these two types may represent the immigration of two tribes from different regions. Or the situation could be more complicated, with any number of immigrations and any population expansion at different times. An important observation from this example: a clade can consist of one or more daughter clades that are much younger than the parent.
This is actually not a surprise as M458 is much younger than the parent clade R1a1a (R1a-M17). Underhill tentatively identifies M458 as a Mesolithic mutation. However, the relevant haplogroup may not have grown much at first, or may have grown and then dwindled over millennia. There appears to have been a recent resurgence from two or more small founder populations. It will be interesting to learn about the culture of these founders.
When it comes to samples from M458, we have more and more of them (Map 10), for example: Fionia (Denmark), Usedom (Germany), Ciepłe (Poland), Lutsk (Ukraine), Kurewanicha (Russia) (The Apricity, 2020) .
On discussion forums such as The Apricity, most of these samples are, for some reason, attributed to the Vikings, not taking into account that the Viking teams included not only Scandinavians, but also Slavs, mainly Pomeranians and Polabians (Polish: wicięgi, Kosiński, 2022a ), as well as the Prusai and the Balts.
New recognitions of M458 are still coming, whether in the Czech Republic or in Hungary (Pannonia). A lot of up-to-date information on this subject can be found not only in publications or scientific articles, which unfortunately quickly become outdated, but also on passionate blogs and thematic discussion forums. For example, in the Anthrogenica forum dedicated to issues related to genetics and anthropology, there are voices with strong arguments in the form of PCA charts (Figure 3), where all medieval L1029 samples are compared with the 6th century early Slavic Av2 sample from Pannonia and with early Germanic samples from Slovakia (DA119) or Germany (average for all early medieval samples from Germany) and there is no indication of their Germanic (rather than early Slavic) origin (137 Ancient Human Genomes, 2020) .
2.4. The Origin of the Indo-Europeans According to the Findings of A. Klyosov
A lot of confusion in the world of archaeogenetics was caused by the research and views of Anatole Klyosov, already mentioned here. He is a Russian professor of biochemistry, a researcher cooperating with the USSR Academy of Sciences, and since 1990 he has been working in the USA, where he has been associated with Harvard University since the 1980s. He was also reportedly the first Internet user in the USSR. He is the creator of the logarithmic method and “DNA genealogy”, as a “new science”, aimed at the synthesis of biology, anthropology, archeology and linguistics, and the implementation of chemical kinetics methods in genetics.
He is also the founder and president of the Academy of DNA Genealogy, under the banner of which he published over 150 issues of the scientific bulletin “Proceedings of the Russian Academy of DNA Genealogy” in the years 2008-2022 with the results of research conducted by this institution and scientists associated with it.
In 2013, Klyosov became the editor-in-chief of the journal “Advances in Anthropology”, published by Scientific Research Publishing (SCIRP). Despite preferring to publish his texts in this magazine, he is also the author of articles in other scientific periodicals (“Nature”, “Journal of Genetic Genealogy”, “Human Genetics”). He has also published many books on the origins of Eurasian peoples and DNA genealogy.
Data from analyzes of available ancient DNA (aDNA) samples, according to Klyosov, already show a fairly clear picture of the migration of the main haplogroups, including R1a/R1b, I1/I2, N, the most common in Europe (Klyosov & Tomezzoli, 2013) .
Klyosov connects hg I with the communities of “Old Europe”, when people lived peacefully, according to our present ideas. He also states that in the Vinča culture (8000 - 5000 ybp), almost all the time of its existence, there were no fortified structures, no weapons were found. Perhaps it was the golden age of “Old Europe”. However, this culture died about 5000 ybp, when the “Old Europe” haplogroups died out, and the Bell Beaker culture (haplogroup R1b) actively populated Europe.
The Eastern European haplotype, one of the hg I subclades, is also called the “Dinaric subclade”, after the name of the Dinaric Alps in the Adriatic. These are the territories with the highest frequency of carriers of this subclade and presumably the region through which the subclade’s bottleneck passes, from where it spread north to the Baltic and to all countries of Eastern Europe, from Poland to Greece, Ukraine, Belarus, Russia (Klyosov, 2012) . Most of them settled in the Balkans. There, after connecting with the carriers of R1a, they gave rise to the ethnos of the southern Slavs, called Illyrians, Thracians, Dacians. haplogroups After several centuries, they increased their numbers and historical activity to such an extent that they created the first Slavic state that was established on the Danube. It is from there that some historians derive the Slavs as such.
About 3000 ybp, carriers of haplogroup N began to move from the Urals to the west, towards the Baltic Sea, and then split into two branches: the South-Baltic and Finnish. The first came to the Baltic coast about 2500 ybp, the second reached the territory of today’s Finland about 500 years later. The northern, Finno-Ugric group, of course, retained its ancient language in its dynamics, while the southern Baltic group met the carriers of the R1a haplogroup, which in the middle of the first millennium BC can already be called Slavs, according to the terminology of Academician V.V. Sedov (1979) .
Currently, the Balts, who linguistically include Lithuanians and Latvians, have the same content of haplogroups R1a and N1c1 (for Lithuanians 38% and 42%, respectively, for Latvians 40% and 38%, respectively). Even among Estonians who are not Balts (Balts belong to the Indo-European language group, and Estonians belong to the Finno-Ugric, Baltic-Finnic branch), the content of haplogroups R1a and N1c1 is also almost the same—32% and 34%. So, according to Klyosov, the Slavs did not assimilate any of them, but only exerted a certain significant genetic and cultural influence on them.
An American biochemist challenges the “Norman theory” about the alleged Ruthenians from Scandinavia, believing that no Normans left any permanent traces in the genetic image of modern Russians. What’s more, based on aDNA data, he claims that the Swedes are closer to the Slavs and Balts than to the Teutons.
Klyosov’s patriotic approach to DNA genealogy shaped not only his view of the cradle of humanity in central Russia, but probably also determined the birthplace of hg R1a there. He believes that this “Ario-Slavic haplogroup” was formed among the Scythians in Siberia, on the current border between Russia and Mongolia (between Altai and Baikal) around 24,000 ybp.
This is argued by the fact that, firstly, the region of South Siberia has been noted many times as a place of carriers of ancient haplogroups. A burial with haplogroup R and an archaeological date of 24 thousand years was found around Lake Baikal. Haplogroup R is the “grandfather” of haplogroup R1a. Further, on the Angara River near Lake Baikal, a burial with haplogroup R1a and an archaeological date of 8000 ybp was found.
Moreover, there are now tribes in northern China where one-third of the population belongs to the R1a haplogroup with an estimated time of common ancestry of 21,000 ± 3000 ybp. This dating was calculated as far back as 2009 (Klyosov, 2009) , when there was even no idea of any snip-based calculations.
The migration map for R1a developed by Klyosov shows that representatives of this haplogroup sat in one place for about 7000 years. Only 15,000 ybp, a group was supposed to move south, reaching Kashmir, and then two millennia later, some of them crossed over to Iran. Between 12 - 11 thousand ybp, R1a appeared in Armenia. Then through Anatolia 10 - 9 thousand ybp, its carriers made their way to the Balkans, to the territory of present-day Bulgaria. 5900 ybp in Pannonia, the southeastern Z-645 clade emerged, which Klyosov calls “Aryan”. 5000 ybp, one branch of Z-280 (South Aryan) reached the area between the Rhine and Elbe (to present-day central Germany), the second one M-458 (West Slavic) over the lower Vistula (Lesser Poland) and Podkarpacie Subcarpathian region < Biała Chrobacja White Chrobatia/Croatia, and the third Z-93 (Eastern Slavic) to Ukraine. Another Pannonian faction passed 500 years later through Odrowiśle area between Oder and Vistula to Scandinavia, giving rise to Z-284 (Scandinavian).
At the same time (4500 ybp), representatives of all three subgroups (Z-280, M-458, Z-93) were said to have made their way to the plains of eastern Russia, and from there spread over the following centuries to the Caucasus (Georgia), Baltic (Latvia), Estonia), to Central Asia, to the Pamir (Tajikistan → Avestan Aryans), Anatolia, Syria, Dacia and Dalmatia, as well as to the east (Southern Urals → Sintashta-Petrovka culture), from where they moved to India 3500 ybp (Vedic Aryans from Sintaszty-Pietrowka) and Iran (from Tajikistan). 3200 ybp, migration to Italy (Venets and Trojans) started from Anatolia, and then the areas of present-day Slovenia and later Noricum, Raetia, were settled. Between 1000-800 BC from the south of what is now Sweden, a group sailed to Britain.
No fossil R1a haplogroups have yet been found in ancient Balkan cultures, but what can you expect when out of 200 burials of the Lepensky Vir culture (Serbia), dated 11,500 - 8000 ybp, only two or three burials have been examined for fossil DNA. However, the positions of the skeletons, where much more data is available, are often typical of haplogroup R1a, i.e. on the side in a crouching position. We are waiting for new data on Balkan haplogroups and sections dated over 8 - 10 thousand ybp.
Also on the issue of language, Klyosov takes a firm stand, claiming that it is one of the ethnic and cultural characteristics, on a par with genes. Before the division of R1a into subgroups, due to migrations in different directions, all of them certainly spoke the language of the native Aryan haplogroup Z645 and used it many centuries later. It is therefore not surprising that, according to the well-known linguist Sergei A. Starostin, modern Russian has 54% of the same basic vocabulary as the old Indian language (Sanskrit). The same applies to other Slavic languages. Therefore, the claim that they “evolved” or “formed” in the middle (or even at the end) of the first millennium AD, as some linguists believe, is, according to Klyosov, completely wrong. Historically, languages do not “evolve” and “form” simultaneously, in their dynamics they develop over millennia, as evidenced by Starostin’s data.
At about 5000 years of age, the Aryan subclade R1a-Z645 split into Aryan branches of the R1a haplogroup, which eventually occupied their territories in Eurasia. A subclade, or branch R1a-M458, formed in its part a group of West Slavic and Central European peoples. The R1a-Z280 branch formed an East Slavic group, largely intersecting geographically with the M458 branch, especially in the west of the East European Plain. The R1a-Z93 branch largely left the East European Plain (its content among the Slavs is now a percentage or fractions of a percent) and moved east, constituting a significant part of the modern male population of Central Asia, Tatarstan, India, Iran, the Middle East and the Caucasian peoples, the region Altai. The same branch was found in Khazarian DNA samples (Klyosov & Faleeva, 2017) . The R1a-Z284 subgroup is currently found in up to 15% - 25% of men in Scandinavia, up to 9% in Scotland, and 3% - 5% in the rest of the British Isles. Slavic peoples practically do not have it.
The most common subclade of R1a-M417 in South Asia today is R1a-Z93, and, realistically, it couldn’t have arrived there earlier than about 2000 BC., so much for the Pleistocene (Map 11).
Since most Scythians with known haplogroups belong to the marker R1a-Z93-Z94-Z2123, it is clear that these Scythians are direct descendants of the southern Aryans. But from the southern Urals, the Scythians did not go south to Hindustan, nor through Tajikistan to the Iranian plateau, nor through the Caucasus to Mesopotamia—Asia Minor, and did not become the common ancestors of the Tatars or Tajiks. The genealogical roots of Tajik DNA go back to the earlier times of the Aryans in the East European Plain 4600 - 4000 ybp, as do parts of the Indians.
The Scythians went to Altai and Mongolia, most of them became nomads and crossed from Altai to the Black Sea region (sometimes further west) and back, spoke mostly Turkic languages (although some believe that they spoke Indo-Iranian or Austronesian languages after all). In the Samara region, fossil DNA of the haplogroup R1a-Z2123 with an archaeological date of 2395-2215 years was found in the remains of the Scythians. The Scythians became the ancestors of most of the Kyrgyz and at least a third of the Karachay-Balkars. Those who were Sarmatians mainly had haplogroup R1b with a series of different subclades. Therefore, after announcing partial results from research on Piast genotypes in Poland, where R1b was found among the Piast elites, Klyosov stated that these are the descendants of those Sarmatians glorified by the Polish nobility, which is not a myth, but a consequence of the coexistence of Slavs and Sarmatians.
It is worth adding here that Underhill announced that Iranian peoples did not settle on the Dniester, Dnieper and Danube, because there are no Iranian R1a-Z93 markers there (Bogdanowicz, 2017) . So if we relate this remark to the Scythians who were supposed to have inhabited these areas in antiquity, it may suggest that they had a different origin, perhaps Eastern European. The name Scythians, known from historiography, in this approach, would therefore apply, similarly to the Germans, to the multi-ethnic population living in a specific geographical area, in these examples—Scythia and Germania.
He also recognizes the Etruscans as Proto-Slavs and Venetians. The Enets (Enetoi, Ἐνετοι) of Paphlagonia, according to ancient historians, became the Illyrian Enets, then the Venetians and Thracians, then apparently the Wends and early Celts of hg R1a who spoke Indo-European languages. Linguists have never found other sources of IE languages for the Celts other than carriers of haplogroup R1a (Klyosov, 2015a) .
The stages of this historical journey of the carriers of the R1a haplogroup, most likely migrants from the Fatyanovo culture, i.e. the ancient Rus, were as follows: migration to Paphlagonia (4300-3500 years), migration to Lydia, as Eneti (at the same time, at the end of this period), migration to Troi, as Eneti (at the end of this period), expulsion of the Eneti-Venet captives in the Apennines, to the future Venice and the Gulf of Venice (3200 years), moving the Veneti north of the Adriatic, rising or joining the Illyrians and Thracians (after 3200 years). Then, in the first half of the 1st millennium BC, i.e. 3000-2500 years, the formation of the early Celts of the R1a haplogroup with Indo-European (IE) languages, followed by the rapid borrowing of the cultural features of the early Celts, the borrowing of their IE languages and their assimilation by the surrounding Europeans of the R1b haplogroup. There was a rapid “erbinization” (adjustment to the digestion of milk protein) of the early Celts, and very quickly by historical standards, over several centuries, Celts in Europe became predominantly carriers of the R1b haplogroup (Klyosov, 2014) .
After all, the history of the migrations discussed here ends with the Norman (Viking) invasions of the British Isles, between 789 and 1066. In them, the haplogroup R1a-Z284 is highly intensified. There was nothing like it in Russia, the Normans did not live on the East European plain. It was the Slavs who interbred all over Europe. But no one wants to admit that they were the initiators of the creation of the most important European civilizations (Klyosov, 2019) .
His entire dating proposal, as well as the routes and time periods of migration of peoples with particular haplogroups, especially hg R1a and R1b, seem to be well grounded in the facts resulting from the analyzes of fossil DNA samples of many scientists to whom Klyosov refers. Where there are ambiguities or lack of data, the American-Russian biochemist leaves room for interpretation, but warns against unfounded confabulations.
Despite this, L. S. Klein (2015) , O. Balanovsky (2014) and his wife E. W. Balanovskaya et al. (2015) , call Klyosov’s views “dangerous genetic demagoguery” and accuse him of practicing pseudoscience (Antonova, 2016) . In response, Klyosov (2018) repeatedly spoke negatively about Balanovsky, who heads the Institute of Genetics of the Russian Academy of Sciences, calling him a “systemic swindler”, and he calls his activities “Balanovshchina” (Russian: Балановщина), adding that these are mainly “innuendos combined with illiteracy” (Klyosov, 2015b) . He also accuses him of unjustified personal attacks, lies and writing nonsense in the field of DNA genetics. Of his wife, who writes critically about Klyos, he writes briefly that “the apple does not fall far from the tree” because together they play the same game, pretending to be academics without having basic knowledge of the topics they talk about.
2.5. Representation of the Slavs in Eurasia According to Genetic Statistics Collected by V. Kubarev
The topic of population genealogy was also taken up by the Russian Valerij V. Kubarev (2011) , doctor of historical sciences and theology, member of the Soviet Academy of Sciences. In one of his articles, he presented a table of the number of Slavs (actually representatives of hg R1a1) in Eurasia, which was developed by his team on the basis of huge research material used to prepare geographical maps of the areas of movement of Slavs, or rather representatives of the R1a1 haplogroup, in Eurasia (Table 2). Based on the given figures from the works of Western scholars, dealing with population genetics, were collected in the tabular form of the number and percentage of the Slavic population among Eurasian countries.
According to data from 2010, 66 million Slavs live in Russia, and about 92 million people together with Ukraine and Belarus. In all of Western Europe, as well as in Russia, there are about 66 million West Slavs. But in Asia, as many as 650 million people have hg R1a1. It turned out that the Slavic haplogroup R1a1 in Eurasia has almost 810 million people, which gives it the second place in the world, after the Chinese haplogroup O (1137 million people for the same year).
Of the total of 806.5 million R1a1 representatives in Eurasia, 100 million are Orthodox, 57.4 million are Catholics and Protestants, 272.2 million are Muslims,
and as many as 376.9 million are Hindus. What is surprising here is the lack of percentage quotas for R1a1 holders who profess Judaism, Buddhism, Taoism or other religions, as well as for natives, atheists, agnostics, etc. It seems that this calculation probably omitted 28 million R1a1 representatives from China and a significant population from both Americas.
In any case, the given numbers indicate that R1a1 cannot be associated only with the Slavs, in the religious and cultural sense, and certainly not with Christianity, which is only in third place after Hinduism and Islam.
Here, of course, the question arises whether all representatives of hg R1a1 can be considered Slavs, or whether we can rather talk about an Indo-Ario-Slavic group. Some commentators claim that all representatives of hg R1a1 and hg I1/I2, as well as people with other haplogroups (J, G, E, R1b, etc.) who speak the Slavic language, are de facto Slavs. It should also be noted that the table provided by Kubarev does not take into account some R1a populations in other countries, for example in Switzerland—1 million people, Saudi Arabia—2 million, Egypt—2 million, Indonesia—15 million, France and England—7 million, etc.
The opposite opinion is held by most academics who believe that genes are not a determinant of ethnicity, nor is language. They give an example of Bulgarians, who are a mixture of different haplotypes, and speak Slavic. However, we can say since they speak Slavic, they are also Slavs, the more so because they consider themselves to be. Another example is the Hungarians, who have a significant share of R1a1 in the population, but speak Magyar and do not feel Slavic or identify with their culture.
As we can see in the table above, less than half of the population of Russia are representatives of R1a1, which we can consider Slavs. The rest are citizens of the Russian Federation with a different ethnic identity. Although many of them speak Russian, they also use their own dialects, profess a different religion and have their own customs, and also feel they belong to a separate family, which is determined by blood kinship. That is why I think that the most accurate determinants of an ethnic group are genes, but also language, culture, customs and one’s own conviction about belonging to a given ethnic group. However, none of these factors can be determinative, but statistics show that language and culture most often follow genes and in the discussion about ethnicities, they are their main distinguishing features.
Kubarev (2011) happens to have quite radical views, not only in political matters—he calls himself the “Grand Prince of All Russia”, but also in historical matters and the origin of Eurasian peoples. He regards Russia as the cradle of the white, Finno-Ugric population, and the Slavs as Asian invaders and tyrants, who are closer to the Mongols, Aryans and Indians than to the Ugro-Finns. Binds them with hg Y-R1a1, but believes that the genotype is given in the male line, and the phenotype—determining the appearance—in the female line, which is not entirely true. Therefore, he argues that it is not surprising that the descendants of the Slavs from Mongolia and Central Asia, as a result of contacts with white, Finno-Ugric women in Russia and Europe (Huns, Avars, Magyars), gained a lighter complexion and began to look like other Old Europeans. In contrast, the Slavs in Central Asia, Inner Mongolia and Hindustan remained ignorant, like the women there.
Of course, Kubarev (2011) does not take into account here the reverse route of migration, namely from Central or even Central and Eastern Europe, to the east and south-east, where representatives of R1a, through contacts with local people with darker complexions or Mongoloid appearance, acquired to some extent its physiognomic features, while their genotype remained unchanged. That’s why we have R1a in the Kyrgyz or Hindu Brahmins. In the latter case, due to the hermetic policy of marrying within a given caste, a large part of them have a lighter complexion than the rest, which I saw myself during my two trips to India.
Also, when I was in Kyrgyzstan, the locals showed me pictures of some Kyrgyz people with typically European features, with fair hair and green or blue eyes, who were supposed to be exterminated by Mongolian and Turkish invaders. Thus, it was the nomadic and expansive peoples who left their genes in Central Asia, and left their mark on the Europoid population, which arrived there much earlier than the steppe hordes of the late Middle Ages and later times.
2.6. Genetic Confirmation of Chang’s View That Everyone in the World Is Related and New Findings on the Origin of Indo-Europeans
In 2013, Peter L. Ralph and Graham Coop (2013) , American scientists in the field of population genetics, published the results of their research, putting forward the theory that everyone in the world is related to each other, which confirmed a similar thesis of Professor Chang. Scientists have found that anyone who lived in Europe 1000 ybp and had offspring is the ancestor of everyone living today and has some European ancestry. The researchers continued their research to show that the common segments came from ancestors who lived 3000 ybp or 100 generations back.
A Polish columnist and enthusiast of history and archaeogenetics, Sławomir Ambroziak (2019) , notes that in the analyzes of these American scientists, for example, “Serbs share more common ancestors with Poles than with Ukrainians, which points to the Vistula basin as the starting point of the Slavic migration”.
The research results of Wolfgang Haak’s team, published in 2015 in “Nature”, were considered groundbreaking. Scholars claimed to have found evidence of a “mass migration” from the Pontic-Caspian steppe to Central Europe, which took place about 4500 ybp. People from the Corded Ware culture of Central Europe (3rd millennium BCE) were found to be genetically closely related to people from the Yamnaya culture of 4700 - 5300 ybp. The authors concluded that their “results support the theory of a steppe origin for at least some of the Indo-European languages of Europe” (Lazaridis et al., 2016; Science Daily, 2015) .
Two other genetic studies in 2015 supported the steppe hypothesis for the Indo-European Urheimat. According to this research, the specific underlays of Y-chromosome haplogroups R1b and R1a, which are found in Yamnaya and other proposed early Indo-European cultures such as Sredny Stog and Chvalyn (Anthony, 2019) , and are now most prevalent in Europe (R1a is also common in South Asia), they were to spread from the Ukrainian and Russian steppes along with the Indo-European languages. These studies also detected an autosomal component present in modern Europeans that was absent in Neolithic Europeans. It was supposed to be introduced with the R1b and R1a paternal lineages as well with Indo-European languages (Mathieson et al., 2018) .
Another Polish enthusiast of population genetics and history, Adrian Leszczyński (2017) , on the basis of data from the research of Haak’s team, notes that in the territories of almost all current countries covered by the Corded Ware culture, fractions of the R1a haplogroup occur in a significant percentage: Poland 57.5%, Belarus 51 %, Russia 46%, Ukraine 44%, Slovakia 41.5%, Latvia 40%, Lithuania 38%, Czech Republic 34%, Estonia 32%, Norway 25.5%, Denmark 16%, Germany 16%, Sweden 16%. Only in Finland and the Netherlands the percentage of occurrence of haplogroups derived from R1a is small and amounts to 5% and 4%, respectively which may confirm that it was the representatives of R1a who were its creators.
He also emphasizes that the “Indo-Iranian” R1a-Z93 and the “Slavic” R1a-Z282 diverged even before the conquest of India. This would apparently mean that it was not the Proto-Slavs who reached India with the Vedas, but, in my opinion, the Vedas (Wends) should be called peoples who still formed a cohesive ethno-genetic-cultural whole, from the period before the separation of these subgroups. And such a split could have occurred in the areas of Central and Eastern Europe, which is not in line with the steppe hypothesis, which, along with the results of the analyzes of subsequent aDNA samples from various parts of Eurasia, began to be criticized.
Taking Haak’s results at face value, even Colin Renrew changed his view on the Anatolian path of the Indo-Europeans, agreeing with Maria Gimbutas. After announcing data that cast doubt on the steppe hypothesis, he made another turn and became a supporter of the theory of continuity from the Neolithic, promoted by the famous Italian linguist Marco Alinei (2003) .
Klyosov, on the other hand, found that in the Yamnaya archaeological culture, all twelve samples of fossil DNA from male bone remains had the same haplogroup R1b (Haak et al., 2015; Allentoft et al., 2015) . Therefore, we should look for the ancestors of the Slavs, in whom hg R1a is dominant, not in Yamnaya, but elsewhere. And the common genetic roots of the Slavs can be seen, apart from the above-mentioned Corded Ware culture, also in Karelia, where on Deer Island (Oleni Ostrov), on the Russian-Finnish border, hg R1a1 was found in the remains of a man who died some 8400 ybp (Fu et al., 2016) . Let’s look at the map from Haak’s publication, where we can see that in present-day Poland there is the greatest genetic compatibility with the genome of this so-called Karelian (Map 12). There were archaeological influences derived from the Świder culture from the late Paleolithic (between 10,600 and 9600 BC) in the area of present-day Poland.
Man from Jelenia Wyspa had haplogroup R1a1 with mutation M459 (Allentoft et al., 2015) . According to Stanisław Pietrzak (2019) , one of the Polish pioneers promoting knowledge in the field of population genetics, he could be an ancestor for Slavs and Poles with the Y-DNA mutation R1a1a-M198, first in the archaeological Janisławice culture and round amphoras, and then with the R1a1a1-M417 mutation in the archaeological Corded Ware culture, and finally also for Polish and Slavic haplogroups with mutations: Z282 and its sons Z280 and PF6155. We find them in the archaeological Lusatian culture. People with the R1a-Z282 mutation live today in the largest number of all countries in the world, because up to 57 percent, among the population of Poland. Also the autosomal genes (about 90% of DNA) of this Karelian show the closest relationship with the autosomes of today’s Poles, as we will see below on the maps.
When we look at the map from the Genetic Atlas showing the occurrence of R1a in Eurasia (Map 13), it is not difficult to see two main clusters with the highest intensity, the first in present-day Poland, and the second in the region of Kashmir (Ladakh) and slightly higher in the vicinity of the Fergana Valley (on the border of Kyrgyzstan, Uzbekistan and Tajikistan).
Such a high concentration of R1a in these regions may indicate a long usucaption of its representatives. However, the question remains whether they came from their cradle in Central Asia and northern India west to the Vistula River (Poland) or the direction of mogration was the opposite from Central Europe to the south-east. I would lean towards the first option.
Looking at the next map, it is impossible not to pay attention to the fact that on the route of the Slavs-Balto-Aryans, i.e. the Vedas/Wends (Veds) to India, they stopped at the bend of the Indus River, where Baltistan (Little Tibet) is now—the country of the Balts, so reminiscent of the Balts (Map 14). But then there is the region of Ladakh (Slavic: “Lady dach”, what means—“the roof of god/godness Lada”, i.e. “God’s peak”), with Leh as its capital, which speaks for itself. They
![]()
Map 13. Modern haplogroup R1a distribution from The Genetic Atlas (Public Domain).
![]()
Map 14. The influence of the Yamnaya steppe culture, according to the Narasimhana team, with my marking in red the area of the potential cradle of Slavic-Balto-Aryans (SBA), from where PIE (actually PSBA—Proto-Slavic-Balto-Aryan) went to Jutland, giving rise to Proto-Germanic languages and to Baltistan on the the Indus and further south to India ( Narasimhan et al. 2019 , modification of the map for illustrative purposes by T.J. Kosiński).
called the mountains the Himalayas, because there is always “hima laja”, meaning in Slavic “winter lies”. And what’s more, the capital of nearby Punjab in Pakistan is also familiar-sounding Lahaur (Lah noble). Not to mention Kashmir (literally: rich country). This appears to be the Veda trail they entered India by bringing the Vedas with them.
However, the situation in archaeogenetics does not stand still and is developing dynamically, and with new data packages, previous hypotheses are verified and new ones appear. For example, Zuzana Hofmanova (2016) in her dissertation analyzed samples of fossil DNA from a burial ground in Serbia. She found that samples from Vlasac, dated to 7400-6200 BCE show similarities to the genotypes of today’s populations from the middle Danube. However, especially populations from northern and eastern Europe—especially Lithuanians—show the highest similarity. Hofmanova did not include the Polish population in her analyses, but taking into account its today’s autosomal proximity to the Lithuanian population, contemporary inhabitants of Poland would be as similar to representatives from Vlasac as Lithuanians. Among the Mesolithic male haplogroups from Vlasac, R1 and I2 were isolated, and R1b was indicated for sample 37. On the other hand, in female haplogroups U5a and U5b prevailed. These are all genes typical of northern European hunter-gatherers since the Paleolithic.
In the course of subsequent research, it turned out that some of the data Haak and his team relied on may be questionable. This may be due to the requirements of high-quality samples necessary for the extraction of male Y-DNA. For example, mentioned by Haak et al. (2015) as a man from R1b from Obłaczkowo (ca. 2800 BC), it was verified by the team of Helena Malmström (2019) as... a woman. Another sample from this site, as R1a, was not questioned. Even older than the samples from Vlasac, 14,000 years old, there is R1b from northern Italy, and R (the quality of the sample did not allow to unambiguously determine whether it is R1, R1a or R1b) from eastern France (about 12 thousand ybp). These data were provided by Mathieson (2018) syndrome. Representatives of the R1 family were certainly earlier in Europe than the carriers of this haplogroup mutation, whose remains were discovered in the Black Sea steppes. Further, in the work of Kendra Sirak’s team (2020) , there was a sample of Y-DNA dated to 3500-3000 BC from northern Romania. This male haplogroup was identified as R1a.
In any case, it can be said that Underhill’s estimates of the early appearance of R1a in Odrowiśle find more and more confirmation in the fossil material.
Views about the Balto-Slavic source from the middle Danube leading to the Vistula and the Dniester (contained in the works of Frederik Kortlandt (2018) , Marco Alinei (2003) and earlier the Russian linguist Oleg Trubachev (1982, 1991) , in IV/III millennium BC, are also supported by the paleogenetic picture. The theses of the team of Wolfgang Haak et al. (2015) about migrations from the steppe, which had an impact on the emergence of European languages, are increasingly undermined. This would mean that on Narasimhan’s map the direction of migration should not be from Yamnaya to the Vistula, but vice versa, from Central Europe to the East.
Russian archaeologist Leo Klein (2017) , proponent of the theory of Norman origins of Russia, antagonist of Klyosov, noted, like this American-Russian biochemist, that the Yamnaya population is dominated by R1b-L23, with Corded Ware men mostly R1a, as far distant R1b clades not found in Yamnaya. Therefore, he claims that the succession from Yamnaya to Corded Ware has no basis in genetic evidence, which he happens to agree with Klyosov. British archaeologist Barry Cunliffe, & Koch (2016) describes this inconsistency as “disturbing to the model as a whole”.
In addition, the team of Oleg Balanovsky et al. (2017) found that most of the Yamnaya genomes studied by Haak and Mathieson belonged to the “eastern” subclade R-GG400 R1b-L23, which is not common in Western Europe, and none belonged to the “western” branch R1b-L51. The authors conclude that Yamnaya could not have been an important source for modern Western European male haplogroups.
In 2020, a new hypothesis was put forward by David Anthony (2020) which aimed to resolve questions regarding the apparent absence of haplogroup R1a in Yamnaya. He speculates that haplogroup R1a must have been present in Yamnaya, but was initially extremely rare, and that the Corded Ware culture is derived from this recalcitrant population that migrated north from the Pontic steppe and greatly expanded in size and influence, later returning to dominance in the Pontic-Caspian steppe.
Recent studies in the field of population genetics have begun to pay attention to mutations that are the result of genetic diseases. Individuals with specific mutations resulting from this type of disease processes could form some fractions of individual haplogroups. An example is the project of Portuguese geneticists.
A group of scientists in the years 1998-2017 conducted research on twenty-one Portugal cystinotic patients were biochemically diagnosed. In addition to trying to find treatments for this disease, researchers have concluded that the most commonly detected cystinosis mutation is a 57-kb deletion that deletes the majority of CTNS gene and in general, is related with high doses of cystine in leukocytes. This deletion is present in almost 50% of all the Northern European and North American origin. However, outside this geographical distribution, the mutation is almost completely absent, especially in the Middle East, therefore migration and/or ethnicity could be the explanation for this fact as well as the concept of possible other founder mutation from those regions (Ferreira et al., 2018) .
The process of identifying minor mutations by geneticists will certainly help to determine even more precisely the origin of individual ethnic groups, their migration routes and timing, as well as mutual relationships.
2.7. Genotypes of Warriors from the Tollense Valley (North-Eastern Germany) from 3300 Years Ago
Although archaeogenetics is an exact science, which limits the field of data manipulation, it is not uncommon for speculations and overinterpretations of research results to occur, and even worse, even the omission of certain information or omissions. This proves that the policy of history still has a great influence on the work of scholars, and the most important thing in all this is simply “our truth”, which is particularly worrying. A sad example may be the discovery of the battle in the valley of Tollence (Polish: Doleńca), called “Pomeranian Troy”, which was described in “Science” as taking place about 3350 ybp, and the found remains contained genotypes similar to contemporary Poles, Scandinavians and peoples of southern Europe (Curry, 2016) .
Until now, however, we are looking forward to a comprehensive report on the study of fossil DNA from this site. A quarter of a century has passed since the discovery of the first traces of the battle, but so far researchers have presented only a few, not very comprehensive studies, instead of a detailed research report (Sell, 2017; Uhlig et al., 2019; Lidke & Lorenz, 2019) . Eventually, as we know, work on the Tollensetal site was discontinued. We can only presume that the research results are not entirely in line with the expected ones, because where did the ancestors of the Poles come from, since with the allochthonous version adopted so far, no Proto-Slavs could have been there. In subsequent messages, instead of Scandinavians, Germans began to appear, Poles and Slavs also disappeared, and there were, for example, Scots. Perhaps the most discussed discovery of recent years still stirs up a lot of excitement, but it seems to put allochthonists up against the wall, which is why all the fuss about delaying the publication of detailed results of the work seems to be a stalling game (Kosiński, 2022b) .
In Christian Sell’s dissertation (2017) , we find some information on the samples from Weltzin, but without detailed commentary on the possible origins of the warriors, or any hypothesis about the causes and results of this battle, as if the author left the subject to historians. The problem is that they are reluctant to use this type of data, and work rather with printed sources, and such sources do not mention anything about this battle, so according to historical methodology, it simply did not happen.
The data provided in several articles published by archaeologists and paleogeneticists studying the remains from the Doleńca (Tollense) region are analyzed in various ways by enthusiasts, giving their interpretations about this battle, as well as the possible lineages of its participants.
The first diagram (Figure 4) shows that the battle in the Tollense Valley was fought between two large groups, Proto-Slavs and Proto-Germans, with a slight participation of other peoples, probably recruited as foreign mercenaries. It can be said that some resemble Finns, Scandinavians and Germans, some Balto-Slavs, and the last part resembles French, Spaniards, Italians. This would confirm the message given in “Science” about the genetic relationship of the warriors from the battle in the Tollense Valley with “southern Europeans and inhabitants of today’s Poland and Scandinavia” (Curry, 2016) .
Some of the PCA charts (Figure 5) show a greater convergence of the samples
![]()
Figure 4. Autosomal DNA distance of the victims of the Battle in the Tollense valley (source: Eurogenes, Wesołowski, 2017a ).
![]()
Figure 5. Convergence of the genomes of Poles (Polish: Polacy) and Germans (Polish: Niemcy) with the warriors from Weltzin (Polish: Wilczyn), according to one of the commentators on the historycy.org website (Domen, 2019) .
from Welzin with the genotype of contemporary Poles, while others believe that they are closer to the genetic profile of Germans (Domen, 2019) . Let us remember that there are more bodies of the losers left on the battlefield, because the victors took their comrades, except perhaps those who fell into the river or swamp.
In my opinion, arguing about whether the warriors from Tollense are closer to the Germans or to the Poles does not make sense. Both of these modern ethnicities, as you can see, have their ancestors there, who were already in Europe in the Bronze Age, or, as scholars date the Battle of Tollense, about 3250 ybp.
And next one (Figure 6), a slightly newer study of the PCA, obtained from the Reich Lab, with a fragment of the area of greatest similarities between the warriors from Wilczyn (German: Welzin) and the inhabitants of Poland, Norway and Central Europe, placed on the website of Carlos Quiles (2019) . Interestingly, there are no similarities to the Scytho-Sarmatian clades here, so these ethnos did not take part in the war.
If one day it is possible to examine all the remains that are still unexcavated there (scholars estimate that about a dozen percent have only been excavated), then it will be possible to make more detailed analyzes and perhaps more credible conclusions, not only who took part in the fighting, but also who won. Of course, one would expect more samples from the remains of the defeated, simply because more of them fell and their corpses remained on the battlefield, while the victors probably took the bodies of their brothers and cremated them. Anyway, the predominance of remains associated with Germanic or Celtic peoples over Balto-Slavic ones may confirm that it was ours who won.
![]()
Figure 6. PCA with marking the area of the smallest distance of genetic similarities of warriors from the Tollense Valley. We can clearly see that in this area there are genotypes of the inhabitants of modern Poland, Norway and Central Europe, and there are significant similarities with representatives of the Unetice culture, Russians, Finns and, on the other hand, Spaniards and Basques (Quiles, 2021) .
It is a pity that we still have an incomplete picture of these many years of research and it is hard to resist the impression that in this particular case access to facts is subsidized and deliberately spread over time, as if waiting for appropriate clothes to be sewn in order to sell them well. To make matters worse, archaeological work at the site has been suspended, apparently due to a lack of funds, which raises additional suspicions.
I guess it’s not only clear to me. Since there is genetic material similar to Poles and peoples related to them, which is confirmed by analyzes and visible on PCA charts, the nonsense hypothesis about the appearance of Slavs in Central Europe only after the fall of Rome should be thrown away.
3. Summary
This review article, of course, does not exhaust the topic of the importance of archaeogenetics as an auxiliary science in the study of the origin of the Slavs. I hope, however, that the presented material makes us realize that “genes do not lie”, it is only necessary to pay attention to whether the results of aDNA analyzes are properly interpreted.
Paleogenetic research shows that the Proto-Slavs could have formed by mixing Old Europeans with hg I (especially I2a) with newcomers with hg R1a. Based on subsequent analyzes of samples of these haplotypes and their distribution on the map of Eurasia, this is highly likely. It is only a question of determining where such a merger of two different ethnicities was to take place, from which one—Proto-Slavic—was created. Everything indicates that it happened in the Balkans, and in addition much earlier, before the migration of R1a carriers to the lands of present-day Iran and northern India.
In the discussion about the origin of the Slavs, genetic arguments are very strong, supported by the views of paleolinguists such as Federik Kortlandt or Marco Alinei, confirming the indigenous theory of the Slavs, long proclaimed by Professor Józef Kostrzewski and a whole group of other scientists from various fields who advocate the indigenousness of the Slavs in Central Europe. The conclusions of aDNA analyzes undermine the false theses from the times of the dominance of “the only correct versions of history” invented in the imperial, papal, Nazi or communist fashion.
Science based on knowledge, without a political background, should serve the purpose of understanding between nations, not quarreling about which of them was or is better or more developed in terms of civilization. It is important, however, that it be based on the truth and not on scholars’ servility towards a specific historical policy.
Acknowledgements
I would like to thank Professor Anatole Klyosov for valuable comments and openness to other views on the ethnogenesis of European peoples, especially the Slavs.