1. Introduction
Physiological responses of plants are stimulated by a great deal of signals from cell-cell and cell-environment interactions, thus leading to a series of signal transductions. These signal transductions are usually accomplished by reversible phosphorylation of protein kinases (PKs). As the substrates and functions of most PKs in plants have not been identified, plant PKs were temporarily named receptor-like kinases (RLKs) [1]. RLKs can be divided into several families according to their extracellular domains. The leucine-rich repeat (LRR) RLKs constitute the largest family of RLKs. Approximately 225 LRR-RLK genes, belonging to 23 subfamilies, have been identified in Arabidopsis. Sixty of them have been described the function [2]. The LRR domain is generally 20 - 30 amino acids long, and forms α/β helix. These structural characteristics allow LRR-RLKs to participate in protein-protein interactions, whereby modulating cellular functions such as signal transductions, stress response, and immunity [1].
In the LRR-RLK XI subfamily, Arabidopsis thaliana (At) HAESA (AtHAE) and two closely related HAESA-LIKE proteins—AtHSL1 and AtHSL2—constitute a small branch [3]. These three homologous proteins have a similar extracellular LRR domain and cytoplasmic Ser/Thr protein kinase domain [3]. A great number of reports have described the function and signaling pathways that AtHAE and AtHSL2 are involved in [3] [4] [5]. AtHAE is specifically expressed in abscission zones of the floral organs as well as at the base of the pedicels and petioles. Disruption of HAE results in delayed abscission of floral organs in Arabidopsis [4]. Single hae or hsl2 mutant appears phenotypically normal, while the double mutant hae/hsl2 has a complete loss of floral organ abscission [6]. INFLORESCENCE-DEFICIENT IN ABSCISSION (IDA) encodes a small secreted peptide that acts as the upstream ligand of HAE and HSL2. Plants with mutations in IDA present with similar phenotypes to hae/hsl2 mutant [6]. AGAMOUS-like 15 (AGL15), a MADS-domain transcription factor, positioned downstream of HAE, creates a positive feedback loop in transcriptional regulation of HAE via a mitogen-activated protein kinase (MAPK) MKK4/MKK5 manner [7]. Subtilisin-like proteinase (SBT) has also been reported to be involved in the IDA-HAE/HSL2 floral organ abscission signaling [8]. Furthermore, IDA-HAE/HSL2 pathway not only functions in developmentally regulated floral organ abscission, but also in drought-induced abscission of cauline leaves [5]. The IDA-HAE/HSL2 signaling is part of a genetic network regulated by auxin and regulates expression of the genes involved in cell wall remodeling during lateral root emergence [9]. Recently, HSL1 has been reported to act as a cognate receptor for CLAVATA3/ESR-RELATED 9/10 (CLE9/10) to regulate stomatal lineage cell division [10]. Genes encoding abscission signaling components of IDA or IDA-LIKE (IDL) peptides and its receptor HSL proteins were also identified in representative Angiosperm orders from an evolutionary prospective [11]. However, HAE and HSL homologues, and their functions in rice have not systematically been validated.
To further understand the functions of HAE and HSL kinases and the regulating networks, we performed a comprehensive genome-wide in silico analysis of the gene family in rice. Our results provide an overview of HAE/HSL family in rice, facilitating further functional studies about this gene family.
2. Methods
2.1. Identification, Chromosomal Location, and Phylogenetic Tree Construction of HAE/HSL Kinase Gene Families
Homologous OsHSL proteins and gene sequences in rice were explored using Arabidopsis HAE, HSL1 and HSL2 with the BLAST suite program in three databases-GenBank at NCBI (https://www.ncbi.nlm.nih.gov/gene), Rice Data (http://www.ricedata.cn/gene), and Ensemble Plants (http://plants.ensembl.org/index.html) [12] —with an inclusion threshold of E < 1e−30. Meanwhile, a phylogenetic tree of the rice LRR-RLK XI family was constructed, and the members that included in the HAEs or HSLs subgroup were also categorized as the homologous OsHSL proteins. The annotations and alternative splicing patterns of the corresponding genes were determined in Rice Data and Ensemble Plants. The exon-intron structures were analyzed using the Gene Structure Display Server. The exact chromosomal locations were determined by BLASTN search of the obtained sequences against Rice Data database and TAIR (The Arabidopsis Information Resource) database (https://www.arabidopsis.org/) and displayed using Map Chartver. 2.2 [13]. An unrooted phylogenetic tree of AtHAE, AtHSLs, and OsHSLs was built by the neighbor joining tree (NJ-tree) facility of this program with bootstrapping (1000 iterations) after manual adjustment of the alignment using the Genedoc program [14].
2.2. Domain Architecture and Protein Structure of HAE and HSL Kinase Family
The transmembrane helices of the proteins were predicted at TMHMM Server ver. 2.0 (http://www.cbs.dtu.dk/services/TMHMM/). LRR domain was predicted using LRRfinder2.0 as described in Offord et al. [15]. Recommended values for upper and lower boundaries are 95% and 80%, respectively. The additional domains (outlier homologues, homologues of known structure), was identified by SMART (http://smart.embl-heidelberg.de/), which contains Swiss-Prot, SP-TrEMBL and stable Ensemble proteomes in Normal SMART. Signal peptides were identified by SIGNALP-5.0. To exhibit the structural divergence of AtHAE, AtHSLs, and OsHSLs, the conserved motifs in the encoded proteins were analyzed with the MEME online program ver. 4.9.0, and visualized with WebLogo [16].
2.3. Co-Expression and Expression Profile Analyses of HAE and HSL Kinases in Rice
The co-expression analysis data was accessed from RiceXPro (http://ricefrend.dna.affrc.go.jp/). Then RiceFREND was used to identify functionally related genes in various biological pathways to construct the correlated gene network [17]. We selected two guide genes, then set “Hierarchy”, and finally clicked “Draw” to generate the network.
The spatio-temporal gene expression profile of rice during its entire development and growth stages, the gene expression profile in the leaves and seeds under various abiotic stress were derived from the microarray data integrated in RiceXPro and GEO databases.
3. Results
3.1. Identification of HAE/HSL Kinase Gene Family in Rice
To identify HAE and HSL kinase family in rice, the amino acid sequences of Arabidopsis HAE, HSL1 and HSL2 were used to perform independent TBLASTN searches through the above-mentioned databases. Thirteen possible HSL1 and HSL2 genes were identified in the rice genome. Next, we clustered the protein kinases in the genome into eleven groups. Altogether 17 possible HSL kinase homologues, including the above-mentioned thirteen sequences, were deemed as putative OsHSLs (Table 1). Of them, only Os11g11890 had been annotated as HSL2, while all the other sequences had been annotated as HSL1 (Table 1).
Most (14/17) of the rice sequences have one splicing pattern; while two sequences (Os01g53920 and Os01g65650) have two patterns, and one (Os06g36270) has five. All three Arabidopsis sequences have only one splicing pattern (Table 1). The rice sequences have different number of introns in different alternative splicing isoforms. Twelve isoforms have one intron, three isoforms have two introns, one (Os11g11890-HSL2) has eleven introns, and one sequence (Os01g53920) does not have any intron (Table 1). Conversely, all three sequences in the Arabidopsis genome have only one intron (Table 1). The three Arabidopsis genes were distributed on chromosomes 1, 4 and 5 (FigureS1(A)). The 17 sequences localized to all the chromosomes in the rice genome, except for chromosomes 3, 4, 7 and 10, with chromosomes 2 and 11 containing most of the sequences (FigureS1(B)).
3.2. Phylogenetic Analysis of HAE/HSL Kinase Gene Family in Rice and Arabidopsis
To illustrate the evolutionary and functional relatedness of HAE/HSL gene families in rice and Arabidopsis, an unrooted phylogenetic tree was constructed with 17 rice and 3 Arabidopsis cDNA sequences. These sequences were found to diverge into three groups (Figure 1). Group Ι and II contain most of the rice sequences. Group III contains only one rice gene (Os11g11890), the only sequence that was annotated as HSL2. Interestingly, three Arabidopsis sequences (AtHSL1, AtHSL2 and AtHAE) constitute a subgroup of group I, which also includes three rice genes. This observation indicates that OsHSLs, AtHAE and AtHSLs probably originated from common ancestral genes [18], and have some functional similarity.
3.3. Domain Architecture of HAE and HSL Kinase Family in Rice and Arabidopsis
Plant RLKs commonly consist of an extracellular LRR domain, transmembrane motif, and intracellular kinase domain. We firstly predicted the transmembrane helices of the HAE and HSL kinases in rice and Arabidopsis. Most OsHSLs have one transmembrane helix except that Os
02
g
13420, Os
02
g
13460 and Os
11
g
12560 do not have any. Although one or more transmembrane helices were also identified at the N-terminal portion of some proteins, such as Os
09 g
15700 and Os
12 g
43660 (FigureS2), these helices are not valid as they are derived from signal peptides [19]. Next, the domain architectures of HAE and HSL kinases of rice and Arabidopsis were analyzed (Figure2). Almost all the proteins have typical LRR-PK domain arrangement, with LRR domain residing at the N-terminus,
![]()
Table 1. Summary of the identified HAESAand HAESA-like kinase family in rice and Arabidopsis.
![]()
Figure 1. Unrooted neighbor-joining phylogenetic tree of OsHSL, AtHAE and AtHSL genes. The phylogenetic tree was generated by MEGA 6 software. Rice MSU IDs and Arabidopsis TAIR IDs are shown on the right side. Groups I, II and III are in blue, pink and red, respectively.
![]()
Figure 2. Diagram of the domain architectures of HAE and HSL kinases identified in rice and Arabidopsis. Red rectangle: signal peptide; Light green rectangle: transmembrane region; Dark green rectangle: LRR; Blue Oval: kinase domain. Arabidopsis TAIR IDs and Rice MSU IDs are shown on the right side of each diagram.
and the kinase domain at the C-terminus. Furthermore, while Os
02g
13420 and Os
11g
12560 seem to have lost their kinase domains during evolution, Os
02 g
13460’s kinase domain is located at the N-terminus. Additionally, Os
11
g
11890 (HSL2) does not have the LRR domain. All the other HAE and HSL kinases have 9-19 LRR repeats, with most members containing 17 repeats. As for AtHAE and AtHSLs, they show all the typical structural properties, such as domain type and arrangement.
3.4. Co-Expression Network of OsHSL Kinase Genes
To assess the function of OsHSL kinases, we performed co-expression analysis of 17 OsHSL genes. Ten genes were found to co-express with other genes (Figure 3). To assess the possible functions of OsHSL kinases, we examined the genes that have been reported to be co-expressed.
Os01g52480 is co-expressed with Os12g43660, whose homologous gene—At4g35985—encodes a senescence/dehydration-associated protein, and is responsive to various abiotic stresses [20]. Os03g27390, co-expressed with Os08g28870, encodes a helix-loop-helix DNA-binding protein, and is likely involved in the regulation of OsChia4a expression in a JA-dependent manner [21]. Os04g45070, co-expressed with Os11g12530, encodes a Remorin (originally named as pp34) and act as signaling scaffolds during development or cellular defense [22].
Os05g45410, co-expressed with Os05g44770, encodes for heat stress transcription factor sp17, whose disruption causes a lesion-mimic phenotype [23]. Os01g14440 is also co-expressed with Os05g44770, and encodes a WRKY transcription factor, and functions in the activation of a defense response against the rice sheath blight fungus and blast fungi [24]. Os07g23710, another gene that co-expressed with Os05g44770, encodes for a cytochrome p450, which is required for anther cutin biosynthesis, and pollen exine formation in rice [25]. Three genes (Os01g23380, Os12g33160, and Os01g15580) are co-expressed with Os01g13800, and they all are NB-ARC domain containing genes, and function in resistance against phytopathogenic fungi and bacteria [26].
![]()
Figure 3. Co-expressed gene networks of OsHSL genes. Peach red, canary yellow, blue, and green circle represent the queried OsHSL genes, gene symbols, transcription factors, and other genes, respectively. Red line segments indicate interactions of the target protein with other proteins, and black line segments indicate possible interactions with other proteins, which may be linked to the target protein. Because the MSU IDs of several genes could not be found, they are shown only by their RAP IDs.
Seven genes (Os01g72170, Os01g37050, Os02g40240, Os04g52190, Os01g0259500, Os04g53860, Os07g36210) are co-expressed with Os02g13510, suggesting that Os02g13510 may be involved in multiple cellular responses. Among them, Os01g72170 is a tau class glutathione S-transferase and confer plants an increased tolerance to salinity and oxidative stresses [27]. Os01g37050 is a ribonuclease III domain protein, which is involved in protein synthesis, RNA interference [28]. Two genes are co-expressed with Os06g36270, such as Os11g12330 encodes a nucleotide-binding site leucine-rich repeat (NBS-LRR) protein, which recognizes specific pathogen-derived products and activates disease resistance signaling pathways [29]. And Os04g03920 encodes a thioredoxin-like domain containing protein. These results indicate that most OsHSLs may function in biotic and abiotic stresses in rice.
3.5. Expression Patterns of HAE and HSL Genes in Rice
The temporal and spatial expression profiles of HAE and HSL genes in rice were investigated (Figure 4). Most OsHSLs showed high expression in the leaf blade, leaf sheath, and root throughout the developmental stages, with Os02g13510 showing the highest expression. Os08g28870 was mainly expressed in the stem, inflorescence, pistil, lemma, and palea, with the anther (1.6 - 2.0 mm) expressing the highest. Os09g15700 also showed high expression in the reproductive tissues, such as ovary at five days after flowering (DAF) and in the embryo stage at seven and ten DAF, with anther (0.7 - 1.0 mm) expressing the highest. Conversely, Os01g65650, Os01g53920, and Os02g13430 showed low expression in almost all the tissues.
To assess if OsHSL kinase genes are related to abiotic stresses, we investigated the induction patterns of OsHSLs in leaves and seeds under various stresses (Figure 5(A)). Nine genes (Os01g13800, Os01g53920, Os02g12910, Os02g13430, Os05g44770, Os06g36270, Os06g36320, Os11g12530 and Os12g43660) were upregulated and three genes (Os02g13510, Os08g28870, and Os09g15700) were downregulated when subjected to salt stress. When under cold stress, most of the genes were downregulated or showed low expression. In the seed, most OsHSLs were downregulated by cold at 4 DAF, with Os09g15700 showing the most downregulation. Six genes (Os01g13800, Os02g13430, Os06g36270, Os06g36320, Os08g28870, and Os11g12530) were upregulated under cold stress at 10 DAF (Figure 5(B)).
4. Discussion
With the completion of the rice genome sequencing project, we have already begun to pay close attention to sequence data mining and interpretation. In this study, we performed a comprehensive genome-wide analysis of HAE/HSL kinase family in rice. Seventeen OsHSL genes were identified in the rice genome. Interestingly, only one of them (Os11g11890) was annotated as HSL2; all the other sequences were annotated as HSL1. None of the sequences was annotated
![]()
Figure 4. Tissue-specific expression patterns of OsHSLs genes. The expression profiles of OsHSL genes in various rice tissues throughout entire growth stages in the field. The numbers that follow the NSU IDs represent the feature numbers, which have been targeted by the corresponding probe. DAF: days after flowering. Data were obtained from the microarray analyses integrated in RiceXPro and normalized based on the mean expression value of each gene in all tissues analyzed. Expression values were again normalized by logarithm with the base of 2 using the R software. Red and blue boxes indicate high and low expression levels for each gene, respectively.
![]()
Figure 5. The expression profiles of OsHSLs in response to abiotic stress. Relative expression profiles of OsHSL genes in leaf (A) and seed (B) in response to drought, salt, and cold are shown as heatmaps. Data were obtained from the microarray analyses integrated in RiceXPro and normalized based on the mean expression value of each gene in all tissues analyzed. Expression values were again normalized by logarithm with the base of two using the R software. Red and blue boxes indicate high and low expression levels for each gene, respectively.
as HAE (Table 1). Phylogenetic analysis showed that OsHSLs, AtHAE and AtHSLs diverged into three groups, with three Arabidopsis sequences constituting a subgroup of group I alongside three rice sequences. Therefore, these homologous genes most likely originated from common ancestral genes and have some functional similarity. Interestingly, several rice sequences that are located on the same chromosomes are clustered together in the phylogenetic tree (Os02g12910, Os02g13460, Os02g13430, Os02g13420, Os02g13510, and Os06g36320, Os06g36270), suggesting that these paralogues were generated via intra-chromosomal duplication events during evolution [30].
Protein sequences and motifs are directly related to protein function. The LRR domain is specifically responsible for binding to signaling molecules to execute signal transduction [31], and promotes receptor-co-receptor complex formation [32], while the kinase domain phosphorylates the OH group of serine or threonine residues of the target protein. Analysis of the conservation and variation in these two domains may suggest functional similarity and divergence in rice and Arabidopsis HAE/HSL families. In this study, we found that Os11g11890 (HSL2) lost the LRR domain during evolution (Figure 1), which may function as cytoplasmic kinase [33]. However, all the other rice HSL kinases have 8 - 18 LRR repeats, with most members containing 17 repeats. These observations indicate the degree of conservation and divergence with respect to ligand perception in the kinase family. Interesting, the LRR repeats of AtHAESA (21) predicted by LRRfinder is less than that observed by crystal structure analysis [32]. The discrepancy between prediction and crystal structure in Mammalian TLR2 was also reported previously [16]. Similarly, Os02g13420 and Os11g12560 lost their kinase domains, while Os02g13460’s kinase domain shifted to the N-terminus, and Os11g11890’s kinase domain duplicated, indicating the degree of functional diversification in these kinases.
Co-expression networks of genes usually reflect the correlation between the expression profiles of different genes and can be suggestive in predicting the gene functions. In this study, we first performed co-expression analysis to predict the function of OsHSL kinases. The results indicate that a large number of the identified genes that are co-expressed participate in multiple biochemical processes, such as biotic and abiotic stresses (Figure 3), suggesting a function of OsHSL gene family in stress responses. Furthermore, the temporal and spatial expression analysis revealed that most OsHSLs exhibited higher expression in the vegetative tissues (such as leaf blade, leaf sheath, and root) than the reproductive tissues, suggesting they function in the growth of rice plant. In contrast, Os08g28870 and Os09g15700 showed higher expression in the reproductive tissues (Figure 4), suggesting they function during the development. Importantly, most OsHSLs changed their expression profiles when subjected to drought and salt stresses (Figure 5(A), Figure 5(B)). As gene expression profiles are somewhat suggestive of physiological processes governed by the genes, these data again suggest that OsHSLs possibly function in biotic stress responses. Until now, the involvement of AtHAE and AtHSL2 in abscission of floral organs has been reported. In addition, AtHAE and AtHSL2 were also involved in mediating stress induced cauline leaf abscission in Arabidopsis under drought stress [5] [34]. No report about the function of OsHSLs exists. For instance, the functions of OsHSLs in abiotic stresses have not been reported previously. Thus our data provide one of the first clues about the functions of the OsHSL gene family.
5. Conclusion
In this study, we performed a comprehensive genome-wide analysis of HAE/HSL kinase families in rice. Seventeen OsHSLs were identified in the rice genome. Of them, only Os11g11890 had been annotated as HSL2, all the other sequences had been annotated as HSL1. Phylogenetic analysis suggested that OsHSLs, AtHAE, and AtHSLs originated from common ancestral genes have some functional similarity. Domain analysis showed that four members lost or evolved their kinase domains, suggesting functional divergence occurred in these kinases. Co-expression analyses and gene expression patterns suggested a possible function of OsHSLs and AtHSL1 in biotic stresses. These results provide a wealth of information to investigators for characterization of the physiological functions and signal transduction pathways governed by OsHSLs.
Acknowledgements
This work was supported by the funding of the Key Program of the Natural Science Foundation of Tianjin (No. 18JCZDJC33700). This work was also supported by the funding of National Natural Science Foundation of China (No. 31870304) and the funding of the Project for Tianjin Rice Industry Technology System Innovation Team (ITTRRS2018005).
Abbreviations
AGL, AGAMOUS-like; CLE, CLAVATA3/ESR-RELATED; DAF, days after flowering; HAE, HAESA; HSL, HAESA LIKE; IDA, INFLORESCENCE-DEFICIENT IN ABSCISSION; LRR, leucine-rice repeat; MAPK, mitogen-activated protein kinase; PKs, protein kinases; RLK, receptor-like kinases; SBT, subtilisin-like proteinase; TAIR, The Arabidopsis Information Resource.
Supplementary Material
Supplementary FigureS1. Distribution of OsHSL, AtHAE, and AtHSL genes on rice and Arabidopsis chromosomes. (A) AtHAE and AtHSL genes are located on 3 of the 5 chromosomes in Arabidopsis genome. (B) OsHSL genes are located on 8 of the 12 chromosomes in rice genome. Chromosomes that do not contain any of the genes are not shown. The constriction on the chromosomes (vertical bars) indicates the position of the centromere. Arabidopsis TAIR IDs and rice MSU IDs are shown on the chromosomes.
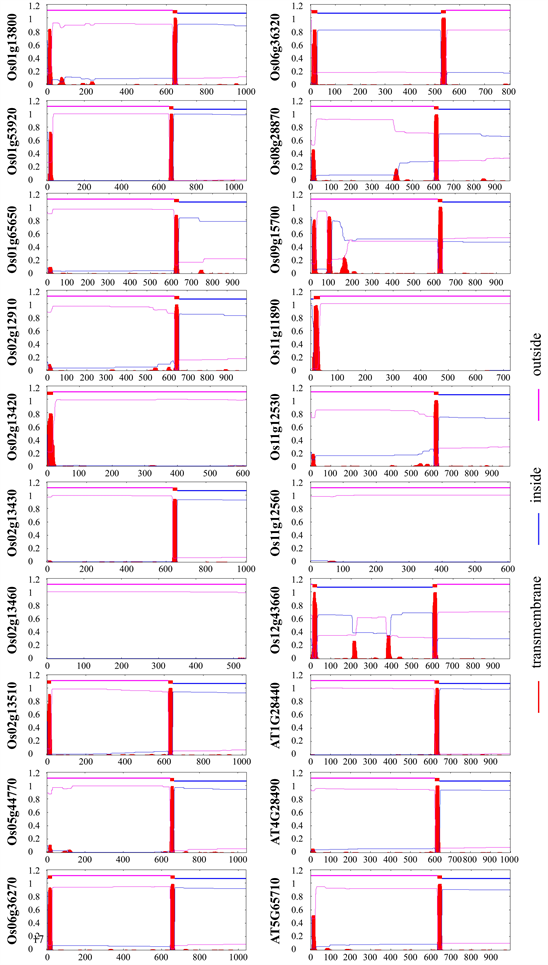
Supplementary FigureS2. Trans-membrane helix (THM) prediction of HAE and HSLs kinase families in rice and Arabidopsis. Amino acids corresponding to red and blue filaments indicate the location in or out of cytomembrane and ones below red grid are considered at trans-membrane region.