ESBL Production and Multidrug Resistance of Salmonella Serovars Isolates in Benue State ()
1. Introduction
One of the most important resistant mechanisms in Gram-negative bacteria against beta-lactam antibiotics is induced by production of beta-lactamases [1]. Extended-spectrum beta-lactamases (ESBLs) have spread threateningly in many parts of the World, and presently comprise over 300 variants.
Extended-spectrum beta-lactamases (ESBLs) are a group of enzymes that have ability to hydrolyze antibiotics, especially third generation cephalosporins and aztreonams but are inhibited by clavulanic acid [2]. These enzymes can be either plasmid or chromosomally mediated, but they are described mainly on plasmid that is frequently found among Enterobacteriaceae.
ESBLs are continuous mutations that change the amino acid configuration near the active site of these β-lactamases, resulting in the development of new enzymes showing extended substrate profiles. ESBLs are prevalent in every part of the world, and in addition, they are found in a meaningful percentage of E. coli and Klebsiella pneumoniae strains. Extended Spectrum beta-lactamases (ESBLs) have been classified into three (3) groups, namely Temoniera (TEM), Sulphydryl group (SHV) and CTX-M with 183, 134 and 103 variants respectively. Paterson et al. [3] reported that among the variants TEM and SHV are the major types in most countries.
Antibiotic resistance, especially to the most commonly used antimicrobials like Chloramphenicol, Ampicillin, and Cotrimoxazole (1) in humans and in animal production systems is of critical concern in Nigeria where multi-drug-resistance (MDR) by Salmonella strains is among the most frequent cause of bacteremia in children (2). Salmonella serotypes with reduced fluoroquinolone susceptibility in humans have been documented (3). The MDR and reduced susceptibility to Ciprofloxacine associated with NTS have been reported from some African countries including Nigeria [4] [5]. The spread of antimicrobial resistance in the food chain is regarded as a major public health issue. This is because of much waste in searching for new drugs and insecurity as the existing drugs will not be effective against the serovars [6].
According to Ohad et al. [7], enteric fever should be treated immediately with antibiotics although widespread resistance to chloramphenicol, ampicillin and cotrimoxazole has been reported. Current treatment of salmonellosis using fluoroquinolones such as ciprofloxacin is under review as the typhoidal serotypes are gradually building strong resistance to the new therapy. New fluoroquinolones such as gatifloxacin and azithromycin hold promise. Multidrug resistance is an increasing problem in S. enterica serotypes. Three types of vaccines against S. typhi are currently available but not licensed vaccine against S. paratyphi A is available. There are vaccines against NTS serotypes Enteritidis and Typhimurium which are effective in poultry but no vaccine available for NTS in human yet. Effectiveness of treatment may depend on the presence of co-infections with other Salmonella species or other organisms such as worms. Precautionary approaches should be adopted. It is advisable that people should not eat foods containing raw eggs or milk such as undercooked fast food. Raw meat should not be cooked in the microwave as it may not reach a high temperature to kill Salmonella or it may be unevenly cooked. Hands must be thoroughly washed after handling reptiles or animal faeces [7].
Salmonella is sometimes antimicrobial resistant which can result in difficulty of treating infections [8]. Because of the impact on human health, zoonotic transmission, and ability to acquire antibiotic resistance (AR), Salmonella has been chosen as the sentinel organism for foodborne disease and for AR monitoring [9]. In the U.S., Salmonella is estimated to cause over one million human infections each year [10]. Most of these infections result in gastroenteritis that resolves after a few days; however, some infections can be chronic or invasive, especially in the very young, the old, and groups of people with compromised immune systems [11]. In these cases, Salmonella infections may require antimicrobial treatment to prevent further morbidity or mortality [11]. First line antibiotic treatment in the U.S. is typically a fluoroquinolone-like ciprofloxacin or a third generation cephalosporin β-lactam such as ceftriaxone, and folic acid pathway inhibitors are also available [12]. However, in children and pregnant women, treatment is usually limited to β-lactams due to fluoroquinolone’s interference with cartilage formation; therefore resistance to β-lactams is a considerable concern in Salmonella [12]. In cases where infection is caused by Salmonella resistant to first line treatments, alternative second line antimicrobials may be used, such as aminoglycosides, or folic acid pathway inhibitors like sulfisoxazole or sulfamethoxazole with or without trimethoprim [13]. In MDR Salmonella infections, the last line treatments are usually the aminoglycoside, amikacin or the carbapenems, imipenem or meropenem which are administered intravenously. Due to observed increases in morbidity and mortality in antimicrobial resistant infections, it has been suggested that resistant Salmonella are more virulent than sensitive strains [14].
Aminoglycoside antimicrobials were first introduced into clinical application in the middle and last half of the twentieth century primarily to treat severe infections caused by Gram-negative bacteria in animals. Their use in treatment of infections in food animals is limited due to both their toxic nature and the persistence of residual antimicrobial in the tissue of the animals. In swine, aminoglycosides including gentamicin, neomycin, or streptomycin have been used to treat intestinal diseases such as scours in weanling pigs and swine dysentery [15] [16]. The synergistic effect of an aminoglycoside antimicrobial with an antimicrobial that targets the cell wall of enterococci such as a β-lactam like ampicillin or penicillin are also used in human medicine to treat enterococcal infections [17]. The aminoglycosides function by binding to the 30S ribosomal subunit inhibiting protein translation.
Salmonella resistance to aminoglycosides is usually an enzymatic modification of the compound; however, in other bacteria, active efflux of the compound or enzymatic modification of the 16S rRNA subunit to prevent the aminoglycoside from binding to its ribosomal target can lead to resistance. Mechanisms of aminoglycoside resistances in U.S. Salmonella animal isolates are primarily due to acetyltransferases, phosphotransferases, and nucleotidyltransferases which modify and inactivate the aminoglycoside [18]. The aminoglycoside acetyltransferases are usually named aac, followed by a numeral in parentheses to designate the target of their enzymatic activity on the aminoglycoside molecule (e.g., aacC3) [18].
Amino glycoside phosphotransferases confer resistance to kanamycin and neomycin, and are usually named aph. These genes also have a designation of the location they modify on the antibiotic (e.g., aph3), and some aph genes also have other names, such as strA and strB which encode resistance to streptomycin. Aminoglycoside nucleotidyltransferases can confer resistance to gentamicin, tobramycin, or streptomycin and include aad and ant groups of genes that can also have extensions to indicate the target of the enzyme. The most common genes reported are variants of aac, aad, aph, and str genes [19] [20] [21].
β-lactams penicillin was one of the first β-lactams developed for clinical use in humans, and was also one of the first antibiotics to which bacteria became resistant. The β-lactams prevent synthesis and maintenance of the peptidoglycan component of the bacterial cell wall by mimicking one of the building blocks used by enzymes to construct peptidoglycan [22]. The β-lactams have a unique four membered “β-lactam” ring that when acted upon by enzymes that build the cell wall, forms an irreversible bond to the enzyme, inactivating it and preventing the enzyme from completing cell wall synthesis. These enzymes are also known as penicillin-binding proteins (PBP). Most resistance to β-lactams is conferred by β-lactamases that enzymatically cleave the β-lactamring and prevent it from bonding to and inactivating cell wall enzymes [22]. Because of this, new β-lactams were synthesized through modification of the chemical groups around the β-lactam ring to produce β-lactams that are resistant to the β-lactamases; other modifications also improved their activity on specific bacteria or accessibility to certain infection sites. These include modified penicillins such as methicillin and oxacillin; the cephalosporins like cephalothin, cefoxitin, ceftriaxone, and cefipime, which are 1st through 4th generation cephalosporins, respectively; and the carbapenems such as imipenem and meropenem [22].
In response to the selective pressure created by these new antibiotics, mutations in β-lactamase genes have also created enzymes that can digest these later generation β-lactams. Some important groups of these are the extended spectrum β-lactamases (ESBLs) [23], cephalosporinases [24], and carbapenemases [25]. However, some β-lactamases can also be inactivated by β-lactamase inhibitors, like clavulanic acid, which bind irreversibly to particular β-lactamases, thus allowing the β-lactam to work when the β-lactam and β-lactamase inhibitor are used as a combined treatment, such as Augmentin (ampicillin/clavulanic acid) [22]. Other resistance mechanisms include genes that encode modified PBPs that have a low affinity for β-lactams and are not inactivated by them or that use different building blocks to construct the cell wall. Efflux of the β-lactam or modification of porins (e.g., ompF and ompC) is also a resistance mechanism to β-lactams. Often these different mechanisms are found in the same bacterium, resulting in high level β-lactam resistance [26]. Most of the β-lactam resistance in Salmonella is encoded by horizontally acquired β-lactamases; however, many other bacteria have an intrinsic β-lactamase, such as ampC found in E. coli [27].
In Salmonella isolated from U.S. animals, the most prevalent β-lactamase genes are blaTEM-1 and blaPSE-1 (a.k.a. blaCARB2) encoding resistance to ampicillin, and blaCMY-2 that encodes resistance to ampicillin, 1st, 2nd, and 3rd generation cephalosporins and is also resistant to β-lactamase inhibitors such as those found in Augmentin [20] [21]. In contrast, other β-lactamases have been detected globally including blaTEM, blaCTX-M, blaIMP, blaVIM, blaKPC, blaSHV, and blaOXA and variants of these genes may encode ESBL or carbapenemase activity [28].
Of particular concern may be the global emergence of the blaNDM-1 metallo-β-lactamase that confers resistance to carbapenems which are often the last line of defense in Gram negative infections [29].
Resistance in Salmonella and other bacteria isolated from animals is often seen by these mechanisms. Mechanisms of phenicol resistance have been floR, cmlA, and cat1. In addition, the chloramphenicol resistance gene floR is often found in the class I integron located in Salmonella Genomic Island 1 (SGI-1) [19] [30].
Quinolones and fluoroquinolones are a synthetic group of antimicrobials used in food animals to combat various infectious agents [30]. Introduced into use over two decades ago, they have broad-spectrum activity coupled with low toxicity and other pharmacokinetic characteristics which make them attractive antimicrobials for use in food animals. A number of fluoroquinolones have been used in food animals including enrofloxacin, difloxacin, marbofloxacin, orbifloxacin, and sarafloxacin [20]. Enrofloxacin and danofloxacin are both useful for treatment of respiratory track disease in cattle; enrofloxacin and sarafloxacin were approved in the mid-1990’s for treatment of chickens and turkeys with E. coli infections [30]. Bacteria resistant to fluoroquinolones used in animals could also be resistant to fluoroquinolones used in human medicine due to the shared mechanism of action of these drugs [30].
Most resistances of Salmonella to these compounds are due to mutations within the genes that encode the enzymes such as gyrA, gyrB, parC, and parE. Most of these mutations occur in the quinolone resistance determining region (QRDR) which is a conserved site in these enzymes targeted by these antimicrobials. Resistance to nalidixic acid and then to fluoroquinolones builds in a stepwise process of mutations in the QRDR region producing an enzyme with a target region that quinolones cannot bind to [31].
Resistance to nalidixic acid and fluoroquinolones like ciprofloxacin has been found in human isolates of bacteria globally. However, animal isolates of Salmonella have very low levels of resistance while the close relative E. coli has higher levels of resistance; for example, only a handful of Salmonella isolated from animals were resistant to ciprofloxacin [32]. Studies have shown that Salmonella resistant to ciprofloxacin also had a growth defect in vitro and in vivo, while E. coli does not [33]. This may be responsible for the low levels of ciprofloxacin resistance seen in Salmonella. Other resistance mechanisms have also been identified including the efflux system, and an aminoglycoside acetyltransferase, aac6 Ib, which can modify and inactivate ciprofloxacin [34].
Interestingly, throughout most of the rest of the world, resistance to 3rd generation cephalosporins in Salmonella is often encoded by ESBLs, which are mutations of several different lineages of β-lactamases [35]. ESBLs are enzymes capable of hydrolyzing extended spectrum/third generation Cephalosporin. These enzymes are also produced by Escherichia coli which enables the pathogen to resist Penicillin, Cephalosporin’s (i.e. First, Second and Third generation), and Monobactams. ESBL Escherichia coli pathogens remain susceptible to Carbapenems and may or may not be susceptible to beta-lactam/beta-lactamase inhibitor combinations [36]. The ESBLs can be found in Salmonella isolated from both animals and humans in Europe, Asia, and South America [37]. Salmonella with ESBLs are infrequently isolated from humans but rarely in food animals [23] [24].
The emergence of Salmonella with antimicrobial resistance is mainly promoted by the use of antibiotics in animal feed to promote the growth of food animals, and in veterinary medicine to treat bacterial infections in those animals [38]. This poses a high risk of zoonotic disease with the transmission of MDR Salmonella strains from animals to humans via the ingestion of food or water contaminated with the animals’ faeces, direct contact or the consumption of infected food animals.
The aim of the study is to determine the prevalence of ESBL-producing and multi-drug resistant Salmonella serovars in Benue State Nigeria.
2. Materials and Methods
2.1. ESBL Confirmation by Double Disc Synergy Test
The method of Nwosu et al. [39], and Nwankwo et al. [40], which conform to Clinical laboratory standards institute (CLSI), was adopted for ESBL confirmation. Isolated colonies were inoculated into peptone water and incubated at 37˚C for 2 - 6 hours. The turbidity was adjusted to 0.5 McFarland’s standard and lawn culture was prepared on Muller-Hinton agar using sterile swab stick. Augmentin (Amoxycillin/clavulanic acid) disc (20/10µg) was placed in the center of the plate, a disc of Cefotaxime (30 µg) and Ceftazidime (30 µg), was placed center to center at a distance of 15 mm to centrally placed Augmentin (Amoxicillin/clavulanic acid) disc. The plates were incubated overnight at 37˚C. Enhanced zone of inhibition between any of the beta-lactam discs and the center disc was recorded. Increased zone of inhibition between any of the third generation cephalosporin antibiotic discs and the Augmentin (Amoxycillin/clavulanic acid) disc relative to the corresponding disc without the beta-lactamase inhibitor was a confirmation of ESBL production.
2.2. PCR Detection of Virulence Genes
All isolates of Salmonella were screened for virulence genes by a Multiplex PCR method as described by Skyberg et al. [41]. Extraction of total genomic DNA from the isolates was done from overnight cultures using DN easy blood and tissue kit (Qiagen, Valencia, CA, USA). The composition of the PCR mixture was as reported by the methods of Skyberg et al. [41]. Table 1 lists all the primers used in molecular characterization of Salmonella. The primers used were as presented in Table 1.
2.3. Agarose Gel Electrophoresis
The extracted plasmids were separated on a 0.8% Agarose gel and electrophoresis was carried out at 80 V for 1 hour 30 minutes. The DNA bands were visualized by Ethidium Bromide staining. Lamda DNA Hind III Marker (Jena Bioscience) was used as DNA molecular weight marker [42].
2.4. Sequencing
All PCR products were purified with Exo sap and sent to Epoch Life science (USA) for Sanger sequencing [43]. The corresponding sequences were identified using the online blast search at http://blast.ncbi.nlm.nih.gov/Blast.cgi.
![]()
Table 1. Virulence gene primers and their nucleotide sequences.
2.5. Sequence Profiling and Identification of Salmonella Strains
The 16s rRNA sequences obtained were compared with known 16s rRNA sequence at National Centre for Biotechnology Information (NCBI) database using Basic Local Alignment Search Tool (BLASTn) algorithm. Identification of the sequences at both genus; species and serovar level was defined as a 16s rRNA sequence similarity at between 95% - 100% with that of the phenotype strain sequence in GenBank. The sequences together with reference sequences derived from the GeneBank were aligned using CLAUSTAL IV [43].
2.6. Antibiotic Susceptibility Test
Antibiotic susceptibility was tested using the Kirby-Bauer disk diffusion method according to the Clinical and Laboratory Standard Institute (CLSI) guidelines. Briefly, pure colonies of bacterial suspension were placed in test tubes and their turbidity adjusted to 0.5 McFarland turbidity standards [44]. The diluted bacterial suspensions were then transferred onto Mueller-Hinton agar plates using a sterile cotton swab and seeded uniformly. Antibiotic impregnated discs were placed on the plate surfaces using sterile forceps. E. coli (ATCC 25922) was used as control. Zones of inhibitions (ZIs) were measured to determine whether the bacteria are susceptible, intermediate or resistant in comparison to Clinical and Laboratory Science Institute critical points [45]. ZIs were classified as: Highly susceptible (≥20 mm); Intermediate (15 - 20 mm) and weakly susceptible (1 - 14 mm).
3. Results
Amplified Plasmids extracted from 18 Salmonella strains are shown in the gel image (Plate 1). The bands made up of 23,130 base pairs. The plasmids contained genes coding for virulence factors and multiple antibiotic resistance. ESBL (Extended-spectrum beta-lactamases) genes were located on the plasmids. Two of the ESBL genes found were TEM genes and CTX-M (415 bp). Strains such as S. enterica Typhimurium-CP014981.1, S. enterica Enteritidis-CP007325.2, S. enterica Typhimurium-CP024619.1, S. enterica Typhimurium-CP023166.1, S. bongori-FR877557 and S. enterica Enteritidis-TY1 possessed TEM genes. S. entericaHeidelberg-CP019176.1, S. enterica Typhi-AL513382.1 and S. enterica Typhimurium-MH196335.1 possessed CTX-M.
Samonella plasmids contained genes that conferred resistance to some antibiotics. Genes such as ParcC (480 bp), GyrA (251 bp), CatA (198 bp) were present (Plate 2). The latter coded for chloramphenicol resistance. InvA plasmid genes (284 bp and 389 bp) typical of virulent Salmonella serovars were identified (Plate 3 and Plate 4). Most of the serovars possessed plasmids that contained TetB gene in the 16sRNA region (571 bp) coding for tetracycline resistance (Plate 5). SitC gene (578 bp) and spvA gene (604 bp) were also identified (Plate 6 and Plate 7).
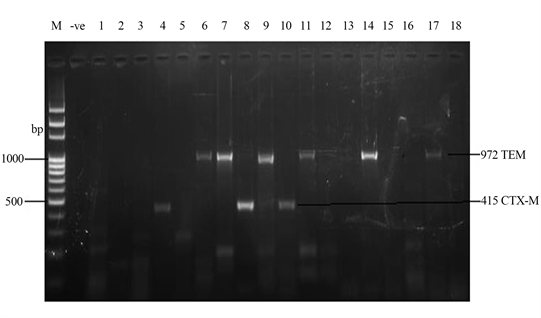
Plate 1. Amplified ESBL Plasmid genes (972 bp and 415 bp) by TEM and CTX-M primers respectively. M = DNA Ladder, −ve = Blank, 1 = S. enterica Agona-392869-2; 2 = S. enterica Paratyphi B-JQ694526.1; 3 = S. enterica Heidelberg-MG663473.1; 4 = S. enterica Heidelberg-CP019176.1; 5 = S. enterica Typhi-AK-1; 6 = S. enterica Typhimurium-CP014981.1; 7 = S. enterica Enteritidis-CP007325.2; 8 = S. enterica Typhi-AL513382.1; 9 = S. enterica Typhimurium-CP024619.1; 10 = S. enterica Typhimurium-MH196335.1; 11 = S. enterica Typhimurium-CP023166.1; 12 = S. enterica Enteritidis-JF951181.1; 13 = S. enterica Huaian-H52.1; 14 = S. bongori-FR877557.1; 15 = S. enterica Typhimurium-LT795114.1; 16 = S. enterica Typhimurium-JQ228518.1; 17 = S. enterica Enteritidis-TY1; 18 = S. enterica Enteritidis-CP018642.1; ESBL = Extended-spectrum beta-lactamases encoded by plasmids.
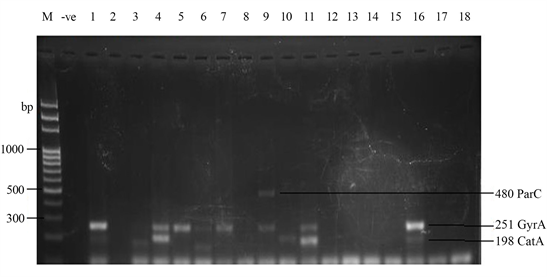
Plate 2. Quinolone Plasmid gene amplified by ParcC (480 bp) and GyrA (251 bp) primers and Chloramphenicol gene amplified by CatA (198 bp) primer. M = DNA Ladder, −ve = Blank. 1 = S. enterica Agona-392869-2; 2 = S. enterica Paratyphi B-JQ694526.1; 3 = S. enterica Heidelberg-MG663473.1; 4 = S. enterica Heidelberg-CP019176.1; 5 = S. enterica Typhi-AK-1; 6 = S. enterica Typhimurium-CP014981.1; 7 = S. enterica Enteritidis-CP007325.2; 8 = S. enterica Typhi-AL513382.1; 9 = S. enterica Typhimurium-CP024619.1; 10 = S. enterica Typhimurium-MH196335.1; 11 = S. enterica Typhimurium-CP023166.1; 12 = S. enterica Enteritidis-JF951181.1; 13 = S. enterica Huaian-H52.1; 14 = S. bongori-FR877557.1; 15 = S. enterica Typhimurium-LT795114.1; 16 = S. enterica Typhimurium-JQ228518.1; 17 = S. enterica Enteritidis-TY1; 18 = S. enterica Enteritidis-CP018642.1.
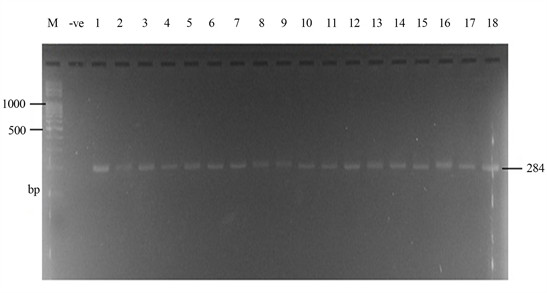
Plate 3. Amplified InvA Plasmid genes (284 bp) by Inv A primer. M = DNA Ladder, −ve = Blank. 1 = S. enterica Agona-392869-2; 2 = S. enterica Paratyphi B-JQ694526.1; 3 = S. enterica Heidelberg-MG663473.1; 4 = S. enterica Heidelberg-CP019176.1; 5 = S. enterica Typhi-AK-1; 6 = S. enterica Typhimurium-CP014981.1; 7 = S. enterica Enteritidis-CP007325.2; 8 = S. enterica Typhi-AL513382.1; 9 = S. enterica Typhimurium-CP024619.1; 10 = S. enterica Typhimurium-MH196335.1; 11 = S. enterica Typhimurium-CP023166.1; 12 = S. enterica Enteritidis-JF951181.1; 13 = S. enterica Huaian-H52.1; 14 = S. bongori-FR877557.1; 15 = S. enterica Typhimurium-LT795114.1; 16 = S. enterica Typhimurium-JQ228518.1; 17 = S. enterica Enteritidis-TY1; 18 = S. enterica Enteritidis-CP018642.1.
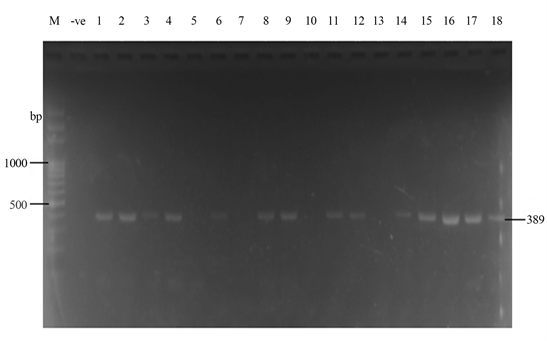
Plate 4. Plasmid amplification of InvA gene (389 bp) by Salm3/Salm4 primers. M = DNA Ladder, −ve = Blank. 1 = S. enterica Agona-392869-2; 2 = S.enterica Paratyphi B-JQ694526.1; 3 = S. enterica Heidelberg-MG663473.1; 4 = S. enterica Heidelberg-CP019176.1; 5 = S. enterica Typhi-AK-1; 6 = S. enterica Typhimurium-CP014981.1; 7 = S. enterica Enteritidis-CP007325.2; 8 = S. enterica Typhi-AL513382.1; 9 = S. enterica Typhimurium-CP024619.1; 10 = S. enterica Typhimurium-MH196335.1; 11 = S. enterica Typhimurium-CP023166.1; 12 = S. enterica Enteritidis-JF951181.1; 13 = S. enterica Huaian-H52.1; 14 = S. bongori-FR877557.1; 15 = S. enterica Typhimurium-LT795114.1; 16 = S. enterica Typhimurium-JQ228518.1; 17 = S. enterica Enteritidis-TY1; 18 = S. enterica Enteritidis-CP018642.1.
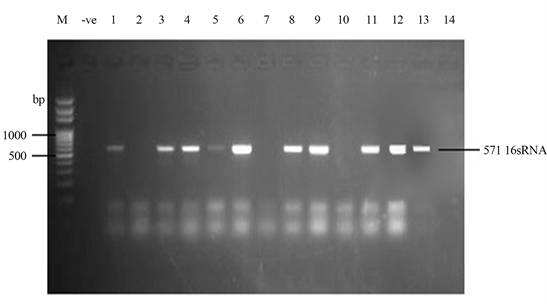
Plate 5. Plamsid amplification of 16sRNA (2) region (571 bp) coding for tetracycline resistance by TET B primer. M = DNA Ladder, −ve = Blank. 1 = S. entericaAgona-392869-2; 2 = S. enterica Paratyphi B-JQ694526. 1; 3 = S. enterica Heidelberg-MG663473.1; 4 = S. enterica Heidelberg-CP019176.1; 5 = S. enterica Typhi-AK-1; 6 = S. enterica Typhimurium-CP014981.1; 7 = S. enterica Enteritidis-CP007325.2; 8 = S. enterica Typhi-AL513382.1; 9 = S. enterica Typhimurium-CP024619.1; 10 = S. enterica Typhimurium-MH196335.1; 11 = S. enterica Typhimurium-CP023166.1; 12 = S. enterica Enteritidis-JF951181.1; 13 = S. enterica Huaian-H52.1; 14 = S. bongori-FR877557.1; 15 = S. enterica Typhimurium-LT795114.1; 16 = S. enterica Typhimurium-JQ228518.1; 17 = S. enterica Enteritidis-TY1; 18 = S. enterica Enteritidis-CP018642.
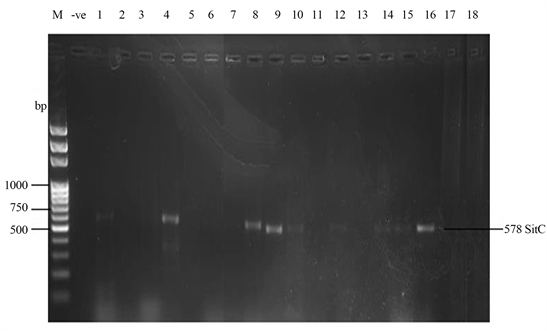
Plate 6. Plasmid amplification of SitC gene (578 bp) by SitC3 primer. M = DNA Ladder, −ve = Blank. 1 = S. entericaAgona-392869-2; 2 = S. enterica Paratyphi B-JQ694526.1; 3 = S. enterica Heidelberg-MG663473.1; 4 = S. enterica Heidelberg-CP019176.1; 5 = S. enterica Typhi-AK-1; 6 = S. enterica Typhimurium-CP014981.1; 7 = S. enterica Enteritidis-CP007325.2; 8 = S. enterica Typhi-AL513382.1; 9 = S. enterica Typhimurium-CP024619.1; 10 = S. enterica Typhimurium-MH196335.1; 11 = S. enterica Typhimurium-CP023166.1; 12 = S. enterica Enteritidis-JF951181.1; 13 = S. enterica Huaian-H52.1; 14 = S. bongori-FR877557.1; 15 = S. enterica Typhimurium-LT795114.1; 16 = S. enterica Typhimurium-JQ228518.1; 17 = S. enterica Enteritidis-TY1; 18 = S. enterica Enteritidis-CP018642.1.
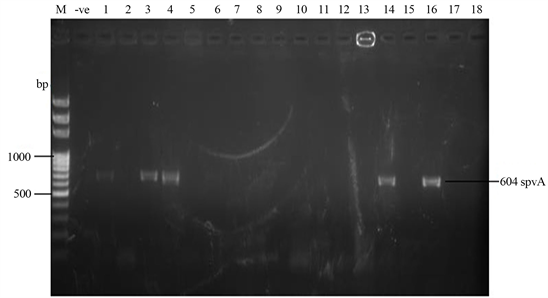
Plate 7. Plasmid amplification of spvA gene (604 bp) by spvA primer. M = DNA Ladder, −ve = Blank. 1 = S. enterica Agona-392869-2; 2 = S. enterica Paratyphi B-JQ694526.1; 3 = S. enterica Heidelberg-MG663473.1; 4 = S. enterica Heidelberg-CP019176.1; 5 = S. enterica Typhi-AK-1; 6 = S. enterica Typhimurium-CP014981.1; 7 = S. enterica Enteritidis-CP007325.2; 8 = S. enterica Typhi-AL513382.1; 9 = S. enterica Typhimurium-CP024619.1; 10 = S. enterica Typhimurium-MH196335.1; 11 = S. enterica Typhimurium-CP023166.1; 12 = S. enterica Enteritidis-JF951181.1; 13 = S. enterica Huaian-H52.1; 14 = S. bongori-FR877557.1; 15 = S. enterica Typhimurium-LT795114.1; 16 = S. enterica Typhimurium-JQ228518.1; 17 = S. enterica Enteritidis-TY1; 18 = S. enterica Enteritidis-CP018642.
Figures 1-7 compare the effects of the various antibiotics tested on the 18 Salmonella strains. Cotrimoxazole (25 µg) gave a high inhibition on S. enterica Enteritidis-TY1 and S. enterica Agona-392869-2 (28 mm) especially after plasmid curing as well as on S. enterica Paratyphi B-JQ694526.1 and S. enterica Heidelberg-MG663473.1 (24 mm) as shown in Figure 1. Cloxacillin (5 µg) had a high inhibitory effect (24 mm) on S. enterica Agona-392869-2 with promising action on S. enterica Paratyphi B-JQ694526.1 and S. enterica Heidelberg-MG663473.1 (20 mm). However, CXC was resisted by many of the Salmonella test strains. After plasmid curing, CXC had an improved inhibition on some test strains (Figure 2).
Tetracyclin (25 µg) gave a moderate inhibition (18 mm) on the two strains of S. enterica Heidelberg, S. enterica Typhimurium-MH196335.1 after plasmid curing and S. enterica Enteritidis-TY1 before curing. Generally, high resistance of many Salmonella strains to the TET antibiotic was observed (Figure 3). Nitrofurantoin (200 µg) showed a high inhibition of 24 mm on S. enterica Paratyphi B-JQ694526.1 and S. enterica Heidelberg-CP019176.1 after plasmid curing. This antibiotic also had inhibitory effect on 22 mm inhibition on S. enterica Enteritidis-TY1 (22 mm) and S. enterica Agona-392869-2 (20 mm). Total resistance to nitrofurantoin was observed in only five strains. Other Salmonella strains displayed weak inhibitory effect of the antibiotic (Figure 4).
Erythromycin (5 µg) was totally resisted by17 Salmonella strains with a weak inhibitory effect on S. enterica Heidelberg-MG663473.1 (Figure 5).
![]()
Figure 1. Comparative inhibition of Salmonella strains by COT. COT = cotrimoxazole (25 µg), Highly susceptibility (≥20 mm), Intermediate susceptibility (15 - 20 mm), Weak susceptibility (1 - 14 mm).
![]()
Figure 2. Comparative inhibition of Salmonella strains by CXC antibiotic. CXC = cloxacillin (5 µg), Highly susceptibility (≥20 mm), Intermediate susceptibility (15 - 20 mm), Weak susceptibility (1 - 14 mm).
![]()
Figure 3. Comparative inhibition of Salmonella strains by TET antibiotic. TET = tetracyclin (25 µg), Highly susceptibility (≥20 mm), Intermediate susceptibility (15 - 20 mm), Weak susceptibility (1 - 14 mm).
![]()
Figure 4. Comparative inhibition of Salmonella strains by NIT antibiotic. NIT = nitrofurantoin (200 µg), Highly susceptibility (≥20 mm), Intermediate susceptibility (15 - 20 mm), Weak susceptibility (1 - 14 mm).
![]()
Figure 5. Comparative inhibition of Salmonella strains by ERY antibiotic. ERY = erythromycin (5 µg), Highly susceptibility (≥20 mm), Intermediate susceptibility (15 - 20 mm), Weak susceptibility (1 - 14 mm).
Gentamicin (10 µg) gave a good susceptibility result across all tests strains. Inhibitory effects ranged from moderate to high level and more pronounced on S. enterica Agona-392869-2, S. Heidelberg-MG663473.1, S. typhi-AL513382.1 and S. typhimurium-CP023166.1. Plasmid curing resulted in higher inhibition of some test organisms by gentamicin although differences are insignificant (T = 1.51, P > 0.05) (Figure 6).
Chloraphenicol (30 µg) displayed high inhibition (22 mm) only on S. enterica Paratyphi B-JQ694526.1. Moderate inhibition was observed on S. enterica Agona-392869-2 (after curing), S. enterica Enteritidis-JF951181.1, S. enterica Huaian-H52.1 and S. enterica Enteritidis-TY1 (Figure 7).
4. Discussion
The results of amplified Plasmids extracted from 18 Salmonella strains are shown in the gel image indicating that the bands were made up of 23,130 base pairs. The ESBL (Extended-spectrum beta-lactamases) genes seen from the study were located on the plasmids, including TEM genes and CTX-M (415 bp). Strains such as S. enterica Typhimurium-CP014981.1, S. enterica Enteritidis-CP007325.2, S. enterica Typhimurium-CP024619.1, S. enterica Typhimurium-CP023166.1, S. bongori-FR877557 and S. enterica Enteritidis-TY1 possessed TEM genes. S. enterica Heidelberg-CP019176.1, S. enterica Typhi-AL513382.1 and S. enterica Typhimurium-MH196335.1 possessed CTX-M.
![]()
Figure 6. Comparative inhibition of Salmonella strains by GEN antibiotic. T (BC and AC) = 1.51, P = 0.141 (P > 0.05), GEN = gentamicin (10 µg), Highly susceptibility (≥20 mm), Intermediate susceptibility (15 - 20 mm), Weak susceptibility (1 - 14 mm).
![]()
Figure 7. Comparative inhibition of Salmonella strains by CHL antibiotic. CHL = Chloraphenicol (30 µg), Highly susceptibility (≥20 mm), Intermediate susceptibility (15 - 20 mm), Weak susceptibility (1 - 14 mm).
The TEM-type ESBLs are derivatives of TEM-1 and TEM-2. TEM-1 was first reported in 1965 from an Escherichia coli isolate from a patient in Athens, Greece, named Temoneira (hence the designation TEM). TEM is further subdivided into three types, namely TEM-1, TEM-2 and TEM-13. TEM-1 is able to hydrolyze ampicillin at a greater rate than carbenicillin, oxacillin, or cephalothin, and has negligible activity against extended-spectrum cephalosporins. It is inhibited by clavulanic acid. TEM-2 has the same hydrolytic profile as TEM-1, but differs from TEM-1 by having a more active native promoter and by a difference in isoelectric point (5.6 compared to 5.4). TEM-13 also had a related hydrolytic contour to TEM-1 and TEM-2 [44].
The name CTX shows the effectiveness of hydrolytic action that β-lactamases exhibit against cefotaxime. Organisms that are known to produce CTX-M-type β-lactamases naturally have cefotaxime MICs in the resistant range (>64 μg/ml), whereas ceftazidime MICs appear habitually prone in that range (2 to 8 μg/ml).
The number of CTX-M-type ESBLs is rapidly intensifying and has now been detected in every populated continent of the world [45]. CTX-M gene has been categorized into five major phylogenetic groups: CTX-M1, CTX-M2, CTX-M8, CTX-M9, and CTX-M25 based on amino acid sequence identity [46]. Most of the CTX-Ms have enhanced their capacity to hydrolyze cefotaxime as compared to ceftazidime. However, several CTX-Ms including CTX-M-15 which is now the most widespread CTX-M enzyme worldwide hydrolyze ceftazidime efficiently [44] [45] [47].
The present studies which undertook appropriate methodologies for detection of Salmonella sp. as described by [48] and antibiotic drug resistance tests as documented in [49] considered eight antibiotics. In this present study, the test drugs exhibited different levels of action and status among Salmonella serovar isolates. This was expected since they are structurally and chemically distinct; each with different pharmacokinetics on the test organisms. Augmentin (AUG) had no effect on all isolates while erythromycin (ERY) inhibited just one out of 18 isolates. Tetracyclin (TET) susceptibility was low but improved to 7 (38.9%) after plasmid was lost mostly at weak level. Chloramphenicol (CHL) susceptibility was also low but improved to 9 (50%) after loss of plasmid but acted in a weak state. Gentamicin (GEN) weakly inhibited all isolates. Cloxacillin (CXC) worked only in double level of action with susceptibility status of 9 (50%) aided by loss of Salmonella plamid. Cotrimoxazole (COT) susceptibility was 12 (66.7%) with triple action level on serovars. Nitrofurantoin (NIT) susceptibility was 13 (72.2%) mostly with triple action level.
The present results are consistent with previous research reports where multidrug resistance was reported on Salmonella serotypes which led to complexity during treatment of infections. Multi-drug resistance and reduced susceptibility to Ciprofloxacin, Fluoroquinolone Chloramphenicol, Ampicillin, and Cotrimoxazole are well documented although with variation in degree of resistance depending on serovar types and locations [3] [50] [51]. In the present study, it is important to strike a balance between antibiotic susceptibility and action level.GEN had the highest percentage susceptibility followed by NIT and COT regardless of presence or absence of bacterial plasmid. Based on level of action, the triple action level of NIT and COT was noted. As reported in Foley and Lynne [26], most pathogenic bacteria possess plasmids as extra circular DNAs that carry genes that code for multidrug resistance especially antibiotic in the host. The emergence of both plasmid mediated antibiotic resistant against conventional antimicrobials and chromosomal resistance has reduced therapeutic options for Salmonella septicaemia in humans. Bacteria can evade the actions of antibiotics using diverse mechanisms. Antibiotic resistance reflects the attack and counterattack of complex microbial flora to an antimicrobial agent in order to establish ecological niches and survive [52]. Salmonella with ESBLs are infrequently isolated from humans but rarely in food animals [19] [23].
To date, there have been reports that the emergence and spread of antimicrobial resistance among zoonotic Salmonella has turn out to be a public health menace [53]. Importantly, Salmonella strains having “clinically important resistance” to some agents like extended-spectrum cephalosporins and fluoroquinolones have been isolated from livestock [54]. In most developing countries, misuse and overuse of antibiotics have contributed to the increasing trend of multi-resistance in Salmonella [55]. In Selangor (center of Peninsular Malaysia), although some reports were found based on the prevalence of Salmonella in different types of foods, but limited information on the surveillance study of Salmonella spp., S. enteritidis and S. typhimurium in beef meat at retail level are available.
The result of our present study showed that more male subjects as compared to female had higher level of multi-drug resistance to the various antibiotics tested when Salmonella plasmid was intact. Loss of plasmid gave equal resistance status to both sexes. This means that the antibiotics were more effective in female subjects against virulent Salmonella seovars. Multi-drug resistance was more pronounced between ages 41 - 50 when Salmonella plasmid was intact or lost. According to WHO [56], persons who are immuno-compromised, children, infants, and the elderly are good candidates of antimicrobial treatment regardless of sex status. Topical research studies have pointed at factors such as the measure of infection, the age as at when the host was infected and their immune response which may also contribute significantly to an efficacious infection [57]. Infections with antimicrobial-resistant strains may concede treatment upshots thereby leading to increased disease and death [56]. In cases where infection is caused by Salmonella resistant to first line treatments, alternative second line antimicrobials may be used [58]. In MDR Salmonella infections, the last line treatments are usually the aminoglycoside, amikacin or the carbapenems, imipenem or meropenem which are administered intravenously [56].
Each of the Salmonella serovar has a unique strain. Virulence gene was found either on the chromosome or on the 16S ribosomal RNA region of the plasmid. Storage capacity of plasmid varied from one strain to another with different nucleotide length and sequence identity. Strains that possessed large storage > 1500 bits are S. paratyphi B strain JQ694526.1 and S. heidelberg strain CP019176.1 (isolated in Gboko). Others include S. bongori strain FR877557.1 isolated from Katsina-Ala and S. enteritidis strain CP018642.1 isolated from Otukpo. The results are also suggestive of the fact that plasmid storage capacity is likely to determine the size of nucleotides. These strains therefore are more likely to carry more virulence genes or more MDR genes on their plasmids.
Some of the Salmonella strains displayed resistance to not more than 3 antibiotics. However, multiple antibiotic resistances (greater than three antibiotics) were highly pronounced in S. bongori strain FR877557.1 (cotrimoxazole, cloxacillin, tetracyclin, augmentin, chloraphenicol and erythromycin); S. typhi strain AL513382.1 resisted all antibiotics except gentamicin; S. typhimurium strain JQ228518.1 (all antibiotics except gentamicin); S. typhimurium strain CP014981.1 (CXC, TET, AUG, NIT and ERY.) and S. typhimurium strain LT795114.1 (CXC, TET, AUG and ERY). Multiple drug resistant salmonella isolates have been reported in previous studies since 1960s and the resistance patterns of Salmonella serovars of public health importance often associated with specific phage types [59]. According to CDCs national antimicrobial resistance monitoring systems, the serovars that have greater resistance to antimicrobials include among others S. typhimurium, S. enteritidis and S. heidelberg which were amongst the serovars isolated from the present study.
The ESBLS are said to be enzymes that are capable of hydrolyzing extended spectrum/third generation cephalosporin which its presence in Salmonella serovars, they are enabled to resist penicillin; First, second and third generation antibiotics and monobactams [36]. Rosangela et al. reported high rates of multi-drug resistance and ESBL production by Salmonella spp. In a similar vein, Maya et al. reported on the emergence of CTX-M-55-type ESBL-producing Salmonella enteric and maintained that the ESBL producing S. enteric strains were the leading cause of human gastroenteritis. Most of the ESBL enzymes were reportedly evolved by bla CTX-M-55 genes and were harbored on conjugative on IncA/C2 plasmids and resistance was reported to up to six (6) additional drug classes and were co-transferred by each plasmid type ESBL-salmonella were reportedly resistant to almost every antibiotic recommended for severe salmonellosis treatment [60]. The emergence of ESBL-producing Salmonella is reported due to selective pressure imposed by the inappropriate use of broad spectrum antibiotics such as the third generation phalospoms [61].
The present research has provided ample information about the range of Salmonella serovars in the six selected health facilities across the six LGAs. It has also provided definite insight on the scenery of antibiotic vulnerability or resistance profile down to the strain echelon of S. serovars in the area under study. The result of the study so provided is critical for the control and management of Salmonella infections in Benue State, North Central Nigeria.
Ethical Consideration
Informed consent was sought from the management of the General Hospitals in the following six Local Government Areas, namely: Katsina-Ala, Kwande, Gboko, Makurdi, Otukpo and Oju respectively through the Executive Secretary, Benue State Hospitals Management Board Makurdi.